Suprachiasmatic nucleus
The suprachiasmatic nucleus (SCN) is a nucleus (an aggregation of nerve cells) in the hypothalamus immediately above the optic chiasm on either side of the third ventricle. The SCN has been described as the 'master clock' of the central nervous system; it generates a circadian rhythm of neuronal and hormonal activities, which regulate many different body functions over a 24-hour period. The SCN contains several cell types and several different peptides (including vasopressin[1] and vasoactive intestinal peptide) and neurotransmitters, and it interacts with many other regions of the brain.
A major function of the SCN in mammals is thought to be to generate biological rhythms with a period of about 24 hours, these 'circadian' rhythms are normally entrained to the light-dark cycle, and they underlie daily rhythms of activity and of hormone secretion. The involvement of the SCN was first shown by experiments in which damage to the SCN in animals resulted in abnormal activity rhythms. Some animals, such as hamsters display very obvious daily rhythms of locomotor activity which can easily be measured by monitoring the time that they spend on a running wheel. In all mammals however many physiological processes are governed by circadian rhythms - for example the secretion of many hormones follows a 24-hour cycle.[2]
Light signals
Neurons in the ventrolateral SCN (vlSCN) have the ability for light-induced gene expression. If light is turned on at night, the vlSCN relays this information throughout the SCN, in a process called entrainment. The SCN receives information about photoperiod via the optic nerve; Melanopsin-containing ganglion cells in the retina have a direct connection to the SCN via the retino-hypothalamic tract. However, neurons in the dorsomedial SCN (dmSCN) can generate a 24-hour rhythm of activity that can persist even in constant darkness (in humans averaging about 24h 11min). The SCN sends information to other hypothalamic nuclei and the pineal gland to modulate body temperature and the production of hormones such as cortisol and melatonin. Rats with damage to the SCN have no circadian rhythms (e.g. they sleep the same total amount, but at random times, for random lengths at a time).
The SCN is one of four nuclei in the brain that receive nerve signals directly from the retina, the other three are the lateral geniculate nucleus (LGN), the superior colliculus, and the pretectum. The LGN passes information about color, contrast, shape, and movement on to the visual cortex and itself signals to the SCN. The superior colliculus controls the movement and orientation of the eyeball. The pretectum controls the size of the pupil.
However a key feature of a true circadian pacemaker, such as the SCN is that it can produce rhythms with an approximate period of 24 hours even in the absence of any change in light. True circadian rhythms thus persist when animals are maintained in constant light or constant dark. Thus light cues do not themselves determine the rhythm, they just "entrain" the rhythm - keeping it locked to the light-dark cycle.
Molecular clockwork
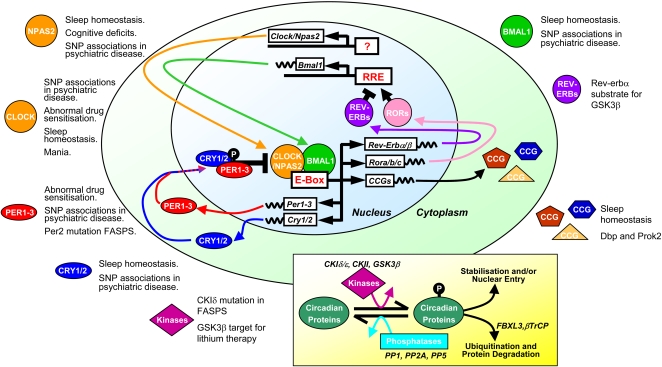
The circadian rhythm in the SCN is generated by a gene expression cycle in individual SCN neurons. This cycle has been well conserved through evolution, and is essentially similar in cells from many widely different organisms that show circadian rhythms. The molecular circadian oscillator incorporates many transcriptional and posttranslational elements, and disruptions in many of these in mice can lead to behavioural disturbances that mirror human neurological and psychiatric disorders (see Figure).
For example, in the fruitfly Drosophila, the molecular clock in neurons is controlled by two interlocked feedback loops. In the first loop, the bHLH transcription factors clock (clk) and cycle (cyc) drive the transcription of their own repressors period (per) and timeless (tim). PER and TIM proteins then accumulate in the cytoplasm, translocate into the nucleus at night, and turn off their own transcription, thereby setting up a 24-hour oscillation of transcription and translation. In the second loop, the transcription factors vrille (vri) and Pdp1 are initiated by CLK/CYC. PDP1 acts positively on Clk transcription and VRI negatively.
These genes encode various transcription factors that regulate the expression of other proteins. The products of clock and cycle, called CLK and CYC, belong to the PAS-containing subfamily of the basic-helix-loop-helix (bHLH) family of transcription factors; these proteins associate to form a heterodimer (CLK-CYC) which in turn initiates the transcription of two other genes, per and tim, whose protein products also dimerize and then inhibit their own expression by disrupting CLK-CYC-mediated transcription. This negative feedback mechanism gives a 24-hour rhythm in the expression of the clock genes. Many genes are suspected to be linked to circadian control by "E-box elements" in their promoters, as CLK-CYC and its homologs bind to these elements.
The 24-hour rhythm could be reset by light via the protein CRYPTOCHROME (CRY), which is involved in the circadian photoreception in Drosophila. CRY associates with TIM in a light-dependent manner that leads to the destruction of TIM. Without the presence of TIM for stabilization, PER is eventually destroyed during the day. As a result, the repression of CLK-CYC is reduced and the whole cycle reinitiates again.
In mammals, circadian clock genes behave in a similar manner. CLOCK (circadian locomotor output cycles kaput) was first cloned in mouse and BMAL-1 (brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1) is the primary homolog of Drosophila CYC. Three homologs of PER (PER1-3) and two CRY homologs (CRY1,2) have been identified . TIM has been identified in mammals, however, its function is still not determined. Recent research suggests that, outside the SCN clock, genes may have other important roles as well, including their influence on the effects of drugs of abuse such as cocaine.[4][5]
Electrophysiology
To regulate other areas of the brain, the neurons in the SCN must communicate circadian information by sending messages in the form of action potentials in a 24-hour rhythm. Such rhythmic changes in electrical activity have been measures in vivo, and even persist when slices of the SCN are maintained in vitro. At mid-day, the firing rate reaches a maximum, and, during the night, it falls again. However, how the gene expression cycle (the so-called 'molecular clock') connects to the neural firing remains unknown.
References
- ↑ Caldwell HK et al. (2008) Vasopressin: behavioral roles of an "original" neuropeptide.Prog Neurobiol 84:1-24. PMID 18053631
- ↑ Hastings MH et al. (2008) Two decades of circadian time. J Neuroendocrinol 20:812-9. PMID 18601704
- ↑ Barnard AR, Nolan PM (2008) When clocks go bad: neurobehavioural consequences of disrupted circadian timing. PLoS Genet 4(5):e1000040 PMID 18516223.
- ↑ Yuferov V et al. (2005). "Biological clock: biological clocks may modulate drug addiction". Eur. J. Hum Genet 13: 1101–3. PMID 16094306.
- ↑ Manev H, Uz T (2006). "Clock genes as a link between addiction and obesity". Eur J. Hum Genet 14: 5. DOI:10.1038/sj.ejhg.5201524. PMID 16288309. Research Blogging.