Halobacterium NRC-1
For the course duration, the article is closed to outside editing. Of course you can always leave comments on the discussion page. The anticipated date of course completion is May 21, 2009. One month after that date at the latest, this notice shall be removed. Besides, many other Citizendium articles welcome your collaboration! |
| Halobacterium sp. NRC-1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
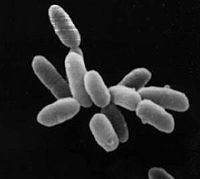 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Halobacterium sp. NRC-1 |
Description and significance
Halobacterium sp. NRC-1 is a halophilic archaea which thrives all over the world in high salt environments, including salt production facilities, brine inclusions in salt crystals, natural lakes and ponds, and salt marshes. Halobacterium sp. NRC-1 is motile using both flagella and gas vesicles, and respond to their environment by moving towards chemicals using a process called chemotaxis and toward or away from light using phototaxis using its sensory rhodopsins. They reproduce via binary fission and grow best in a 42 degree Celsius aerobic high salt environment.
Halobacterium sp. NRC-1 is very easy to culture in the lab, and is genetically tractable. It's genome has been completely mapped and whole-genome DNA microarrays are available to investigate gene expression. This makes it an excellent model microorganism for research into the basic cellular process and gene expression as well as for teaching.
Genome structure
The genome of Halobacterium sp. NRC-1 was published in 2000. Since that time, a combination of genetic, transcriptomic, proteomic and bioinformatic approaches have provided insights into both its extremophilic lifestyle as well as fundamental cellular processes common to all life forms.[1]
Halobacterium sp. NRC-1 contains the smallest genome to date among the halophiles. It is 2,571,010 bp in size, and is composed of a large GC-rich chromosome (2,014,239 bp, 68 % G+C), and two smaller extrachromosomal replicons, pNRC100 (191,346 bp) and pNRC200 (365,425 bp), with 58–59 % G+C composition. The two smaller replicons contain 145,428 bp of identical DNA and 33–39 kb inverted repeats catalyzing inversion isomers, and the majority of the 91 IS elements, representing 12 families, found in the genome. As a result of the large number of repeated sequences, genome assembly required extensive genomic mapping and an ordered clone library of pNRC100. Of the 2,630 likely protein-coding genes in the genome, 2,532 are unique. Halobacterium predicted proteins were found to be highly acidic [27] and a substantial number had bacterial homologs as their closest relatives, suggesting that they might have been acquired through lateral gene transfer. In addition, 52 RNA genes were also identified; however, the 16S rRNA sequence and other unique characteristics did not allow placement within a validly described Halobacterium species, and this point has been the subject of some controversy. Interestingly, about 40 genes in pNRC100 and pNRC200 code for functions likely to be essential or important for cell viability (e.g. thioredoxin and thioredoxin reductase, a cytochrome oxidase, a DNA polymerase, multiple TATA-binding proteins (TBP) and transcription factor B (TFB) transcription factors, and the only arginyl-tRNA synthetase in the genome). As a result, these replicons were suggested to be essential "minichromosomes" rather than megaplasmids.[1]
Cell structure and metabolism
Halobacterium species are obligately halophilic microorganisms that have adapted to optimal growth under conditions of extremely high salinity—10 times that of sea water. They contain a correspondingly high concentration of salts internally and exhibit a variety of unusual and unique molecular characteristics. [2]
Ecology
Pseudomonas putida has an incomparable effect on the environment. They are able to protect plants from pests, promote plant growth, and clean up organic pollutants found in soil and water.
The surface of the root and the soil that surrounds it are loaded with nutrients released by the plant. This environment is optimal for microbial growth. Pseudomonas putida is attracted to this area, and in turn promotes plant growth and even protects the root against pathogens. Two key elements that allow P. putida to attach in the first place is that they are motile and chemotactic towards the root output. After the initial attraction and migration toward the root, the bacteria immediately begins to grow and divide, forming multiple colonies around the root. The maximum population size is directly related to root weight, and once it is reached the number of colonies will stay constant. All of this can happen in less than 48 hours!
Pseudomonas putida play a huge role in bioremediation, or the removal or naturalization of soil or water contaminants. They can degrade toluene, xylene, and benzene, which are all toxic components of gasoline that leak into the soil by accidental spills. Other strains can convert styrene, better known as packing peanuts, which do not degrade naturally, into the biodegradable plastic polyhydroxyalkanoate (PHA). Methods used to get rid of styrene include incinerating it, spreading it on land, and injecting it underground, all of which release the toxins into the environment. Styrene can cause muscle weakness, lung irritation, and may even effect the brain and nervous system. Due to the fact that P. putida can use styrene as its only source of carbon and energy, it can completely remove this toxic chemical. P. putida can also turn Atrizine, an herbicide that is toxic to wildlife, into carbon dioxide and water.
Application to Biotechnology
Pseudomonas putida is being used in conjunction with Escherichia coli for developing new drugs. This study focuses on myxochromide S, a compound produced by Stigmatella aurantiaca, but the method is revolutionary in that there is unprecedented expression of gene clusters. The beginnings of many new drugs are from natural sources, such as plants and microorganisms, but they are too expensive to harvest from the origin. Combinatorial biosynthesis has revolutionized drug development by allowing the structure of certain molecules to be changed within an organism. With this metabolic engineering, where genes are introduced and their expressions are tightly controlled, successful production of drugs is possible. Pseudomonas putida is unique in that it allows the expression of a large biosynthetic cluster, producing five times as much myxochromide S as Stigmatella aurantiaca. This will also permit scientists to connect multiple clusters of genes onto a single DNA fragment. [3]
Pseudomonas putida is able to purify fuel, a capability that the petroleum industry has taken great interest in. As previously mentioned, P. putida is able to convert styrene, a toxic waste product, into a biodegradable plastic. The strain CA-3 turns styrene into a stored energy source, in the form of a plastic polymer called polyhydroxyalkanoate (PHA). Using styrene its only source of carbon and energy, the styrene is completely used up, creating an elastic type of polymer. This polymer can then be used in the production of drug carriers, plastic coating of cardboard, and medical implants.
Current Research
"Benzene, Toluene, and Xylene Biodegradation by Pseudomonas putida CCMI 852"
Gasoline spills create a large amount of toxic pollution in the environment, being that the major components are benzene, toluene and xylene isomers. Catalogued by the U.S Environmental Protection Agency as “priority pollutants”, gasoline is a main cause of water well and spring contamination. Pseudomonas putida can successfully degrade these dangerous components of gasoline, and can aide in the cleanup of such pollutants. This article discusses the research being done in order to determine what environment and mixture of compounds will allow for the most degradation. The metabolic pathway that P. putida uses to break down these compounds is investigated. The TOL pathway does not utilize benzene as a substrate, while the TOD pathway does. Various combinations of these elements of gasoline were used during experimentation. Maximum degradation occurred when each compound was alone in solution. Once any other compound was introduced, the rate automatically dropped, but it dropped most drastically when benzene was introduced. Toluene was degraded at a rate twice as fast as xylene. Benzene concentration always remained unchanged, when alone or in a mixture. It is then obvious that P. putida did not degrade benzene, even when no other compounds were present. It is suggested that this P. putida CCMI 852 strain contains a TOL plasmid, therefore preventing the degradation of benzene. [4]
"Diversity and activity of biosurfactant-producing Pseudomonas in the rhizosphere of black pepper in Vietnam"
Black pepper, a major crop and source of income for the country of Vietnam, is also the most important spice crop in the world. This ‘King of Spices’ is particularly susceptible to the pathogen Phytophthora capsici, which eats away at the roots of the plant and causes death and disease. It can potentially cause up to 40-50% of crop death, and in Vietnam an annual loss of around 20%. Pseudomonas putida was found to produce biosurfactants, which disrupt the membrane of the Phy. Capsici zoospores, causing death within minutes. Using P. putida as a method for controlling Phy. Capsici will be more effective than using pure biosurfactants created in a lab. One reason for this is that the chemical form may not be delivered efficiently to the roots, because they must fully penetrate the soil to the level of the rhizosphere. P. putida is chemotactic towards root output, and move via flagella directly toward the root, subsequently creating secure colonies. Using P. putida as a pesticide will also be a long-lasting form of treatment, because the colonies not will get washed away by a rainstorm like the chemical form could.
Rhizosphere samples were taken from three different districts in Vietnam, and various testing was done with different biosurfactanct producing organisms and bacteria. P. putida provided a significant amount of prevention of plant wilt, providing adequate protection from Phy. capsici. This was greater then when contrasted with other bacteria tested, such as Bacillus spp., Trichoderma harzianum, and P. flourescens. When P. putida was introduced to plant root when no pathogen was present, there were more interesting results. This bacterium had considerably increased shoot height and weight, and also raised the number of roots grown. More research is being done in the area, as it further development of this idea will help control plant pathogens. [5]
"Accumulation of Polyhydroxyalkanoate from Styrene and Phenylacetic Acid by Pseudomonas putida CA-3"
Pseudomonas putida has the unique ability to transform a toxic pollutant into a biodegradable plastic. Over 25 million kilograms of styrene, a compound that can cause respiratory tract infection, muscle weakness, and narcosis, is released into the environment every year in the U.S. alone. The product, polyhydroxyalkanoate, can be used as synthons for certain antibiotics, vitamins, and anti-cancer drugs. They are also used in other areas of medical applications such as tissue engineering and wound management. The pathway and mechanism for this transformation is investigated, where aromatic hydrocarbons are transformed into aliphatic PHA. During the growth cycle, there is the strict use of carbon and nitrogen with styrene, glucose, and phenylacetic acid by P. putida as the carbon and energy source. The effects of altering the carbon to nitrogen ratio and level are tested in relation to the accumulation of the product PHA.
The study showed that when nitrogen levels dropped, PHA production began. The effect of carbon supply was studied as well, and only when carbon to nitrogen ratios of 9:1 for cells on glucose, 10:1 for cells on phenylacetic acid, and 14:1 for cells grown on styrene were reached did PHA begin to accumulate. Next, nitrogen was manipulated while carbon was held constant, showing that the highest PHA content was found when nitrogen concentration was the lowest.
The manner in which styrene is converted to PHA is through fatty acid de novo biosynthesis, which is catalyzed by 3-hydroxy-acyl-ACP:CoA transacylase. The plastic polymer has a destruction point of 265 degrees Celcius, and a uniquely low molecular weight and high polydispersity. This is also the first time that an aromatic substrate is converted into aliphatic PHA. Further research will be done to investigate ways to increase PHA yield from styrene using P. putida.[6]
References
- ↑ 1.0 1.1 DasSarma S et al. (2006), "Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1", Saline Systems 2 (3), DOI:10.1186/1746-1448-2-3
- ↑ Ng, Wailap Victor, et al. (2000-10-24). "Genome sequence of Halobacterium species NRC-1". Proceedings of the National Academy of Sciences of the United States of America 97 (22): 12176-12181. DOI:- 97 VL - 97. Retrieved on 2009-04-18. - 97 Research Blogging.
- ↑ Bechthold, A. “Exploiting Pseudomonas putida for Drug Development”. Chemistry & Biology. March, 2005. Vol. 12, Issue 3. p. 261
- ↑ Otenio, M.H., Da Silva, M.T.L., Marques, M.L.O, Roseiro, J.C., Bidoia, E.D. “Benzene, Toluene, and Xylene Biodegradation by Pseudomonas putida CCMI 852”. Brazilian Journal of Microbiology. 2005. P. 258-261.
- ↑ Tran, H., Kruijt, M., Raaijmakers, J.M. “Diversity and Activity of Biosurfactant-producing Pseudomonas in the Rhizosphere of Black Pepper in Vietnam”. Journal of Applied Microbiology. March, 2008. Vol. 104. p. 839-851.
- ↑ Ward, Patrick G., de Roo, Guy, O’Connor, Kevin E. “Accumulation of Polyhydroxyalkanoate from Styrene and Phenylacetic Acid by Pseudomonas putida CA-3”. Applied and Environmental Microbiology. April, 2005. Vol. 71. p. 2046-2052.
[7]↑DNA isolation, from Qiagen
[8]↑[1]