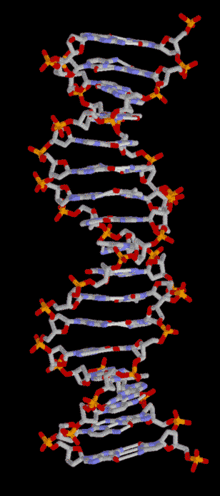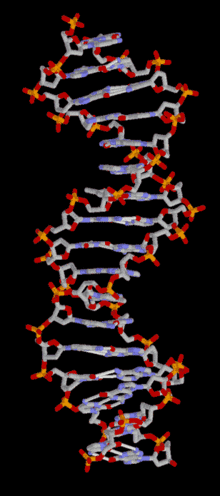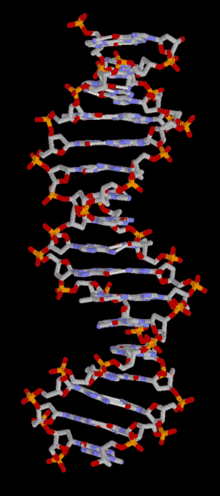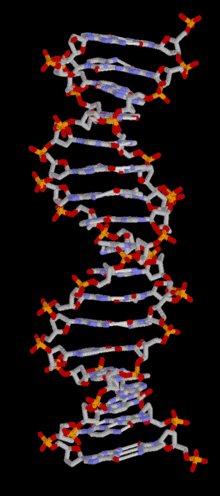
DNA
Deoxyribonucleic acid (DNA) is a very large biological molecule found in almost every cell and is responsible for providing information required for the development and reproduction of living things. Every living organism has its own unique DNA sequence (code) similar to a 'barcode' or 'fingerprint'. This code, called a genome, is used primarily to control the synthesis of proteins. There are two important genetic features of DNA for life. The most important feature is the complementary duplex structure, often referred to as the "double-helix", that enables the accurate replication of DNA.[1] Second, the internally coded, inheritible information defines the genotype which provides the necessary instructions to produce the phenotype, or outward physical manifestation an organism.
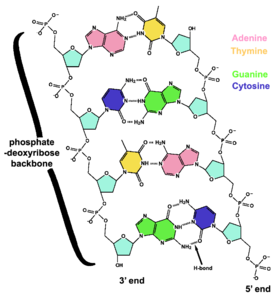
DNA is a long polymer comprised of simple units called nucleotides which are held together by a sugar phosphate backbone (called a nucleoside). Attached to each sugar molecule (deoxyribose) is a molecule of one of four bases; Adenine (A), Thymine (T), Guanine (G) or Cytosine (C). Each base is complemented by the opposing base. Because of the chemical and physical structure of these bases, adenine always pairs with thymine and guanine always pairs with cytosine. These pairs are identical in size and shape and will fit between the backbones in any configuration. Each base pair can have one of four configurations TA, AT, GC and CG. It is the sequence of at least three of these base pairs that form a gene on the DNA strand that encodes the information. This complementarity forms the basis of semi-conservative DNA replication — making it possible for DNA to be copied relatively easily, while accurately preserving its information content.
Template:TOC-left In most organisms, DNA is in a double-helix formation consisting of two DNA strands coiled around each other, in a head-to-tail "antiparallel" orientation, held together by hydrogen bonds between bases on opposing strands. The entire DNA sequence of an organism is called its genome. In animals and plants, most DNA is stored inside the cell nucleus. In bacteria, there is no nuclear membrane around the DNA, which is in a region called the nucleoid. Some organelles in eukaryotic cells (mitochondria and chloroplasts) have their own DNA with a similar organisation to bacterial DNA. Viruses have a single type of nucleic acid, either DNA or RNA, directly encased in a protein coat called the capsid.
Overview of biological functions
DNA contains the genetic information that is the basis for living functions including growth, reproduction and evolution. This information is held in segments of the DNA called genes that may span in size from a minimum of three DNA base-pairs to many thousands of base-pairs. In eukaryotes (organisms such as plants, yeasts and animals whose cells have a nucleus) DNA usually occurs as several large, linear chromosomes, each of which may contain hundreds or thousands of genes. Prokaryotes (organisms such as common bacteria) generally have a single large circular chromosome, but they often possess other miniature chromosomes called plasmids. The set of chromosomes in a cell makes up its genome; the human genome has about three billion base pairs of DNA arranged into 46 chromosomes [2] and contains 20-25,000 genes[3].
There are many interactions that happen between DNA and other molecules to coordinate its functions. When cells divide, the genetic information must be duplicated to produce two complementary copies of DNA in a process called DNA replication. When a cell uses the information in a gene, the DNA sequence is copied into a complementary single strand of RNA in a process called transcription. Of the transcribed sequences, some are used to directly make a matching protein sequence by a process called translation (meaning translation from a nucleic acid polymer to an amino acid polymer). The other transcribed RNA sequences may have regulatory, structural or catalytic roles.
Replication
- Further information: Replication of a circular bacterial chromosome
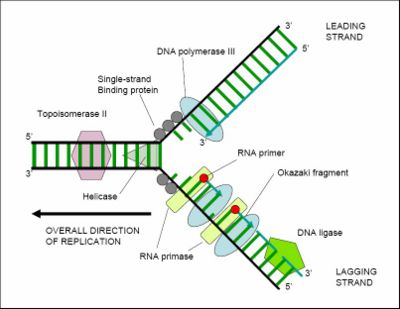
For an organism to grow, its cells must multiply, and this occurs by cell division: one cell splits to create two new 'daughter' cells. When a cell divides it must replicate its DNA so that the two daughter cells have the same genetic information as the parent cell. The double-stranded structure of DNA provides a simple mechanism for this replication. The two strands of DNA separate, and then each strand's complementary DNA sequence is recreated by an enzyme called DNA polymerase. This makes the complementary strand by finding the correct base (through complementary base pairing), and bonding it to the original strand. DNA polymerases can only extend a DNA strand in a 5' to 3' direction, so other mechanisms are needed to copy the antiparallel strands of the double helix.[4] In this way, the base on the old strand dictates which base appears on the new strand, and the cell (usually) ends up with a perfect copy of its DNA. However, occasionally mistakes (called mutations) occur, contributing to the genetic variation that is the raw material for evolutionary change.
DNA replication requires a complex set of proteins, each dedicated to one of several different tasks needed to replicate this large molecule in an orderly and precise fashion. Further capacity for DNA replication with substantial rearrangement (which has major implications for understanding mechanisms of molecular evolution, is provided by mechanisms for DNA movement, inversion, and duplication, illustrated by various mobile DNAs such as transposons and proviruses.
Transcription and translation
Within most (but not all) genes, the nucleotides define a messenger RNA (mRNA) which in turn defines one or more protein sequences. This is possible because of the genetic code which translates the nucleic acid sequence to an amino-acid sequence. The genetic code consists of three-letter 'words' called codons formed from a sequence of three nucleotides. For example, the sequences ACU, CAG and UUU in mRNA are translated to threonine, glutamine and phenylalanine, respectively.
In transcription, the codons are copied into mRNA by RNA polymerase. This copy is then decoded by a ribosome that reads the RNA sequence by base-pairing the mRNA to a specific aminoacyl-tRNA; an amino acid carried by a transfer RNA (tRNA). As there are four bases in three-letter combinations, there are 64 possible codons, and these encode the twenty standard amino acids. Most amino acids, therefore, have more than one possible codon (for example, ACU and ACA both code for threonine). There is one start codon (AUG) that also encodes for methionine) and three 'stop' or 'nonsense' codons (UAA, UGA and UAG) that signify the end of the coding region.
Genes
Our understanding of the various ways in which genes play a role in cells has been continually revised throughout the history of genetics, starting from abstract concepts of inheritable particles whose composition was unknown. This has led to several modern different definitions of a gene. One of the most straightforward ways to define a gene is simply as a segment of DNA that is transcribed into RNA - that is - the gene is a unit of transcription. This definition encompasses genes for non-translated RNAs, such as ribosomal RNA (rRNA) and transfer RNA (tRNA), as well as messenger RNA (mRNA) which is used for encoding the sequences of proteins.
A second approach is to define a gene as a region of DNA that encodes a single polypeptide. By this definition, any particular mRNA transcription unit can cover more than one gene, and thus a mRNA can carry regions encoding one or more polypeptides. Such a multi-genic transcription unit is called an operon.
Other definitions include consideration of genes as units of biological function. This definition can include sites on DNA that are not transcribed, such as DNA sites at which regulatory and catalytically active proteins concerned with gene regulation and expression are located. Examples of such sites (loci, sing. locus) are promoters and operators. (Locus is a genetic term very similar in meaning to gene, and which refers to a site or region on a chromosome concerned with a particular function or trait.)
All of the cells in our body contain essentially the same DNA, with a few exceptions; red blood cells for example do not have a nucleus and contain no DNA. However, although two cells may carry identical DNA, this does not make them identical, because the two cells may have different patterns of gene expression; only some genes will be active in each cell, and the level of activity varies between cells, and this is what makes different cell types different. The "level" of gene expression (for a given gene) is used sometimes to refer to the amount of mRNA made by the cell, and sometimes to refer to the amount of protein produced.
All humans have essentially the same genes, but have slightly different DNA; on average, the DNA of two individuals differs at about three million bases. These differences are very rarely in the protein-coding sequences of genes, but some affect how particular genes are regulated — they may affect exactly where in the body a gene is expressed, how intensely it is expressed, or how expression is regulated by other genes or by environmental factors; these slight differences help to make every human being unique. By comparison, the genome of our closest living relative, the chimpanzee, differs from the human genome at about 30 million bases. [5]
Genomes
In eukaryotes, DNA is located mainly in the cell nucleus (there are also small amounts in mitochondria and chloroplasts). In prokaryotes, the DNA is in an irregularly shaped body in the cytoplasm called the nucleoid.[6] The DNA is usually in linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. The human genome has about three billion base pairs of DNA arranged into 46 chromosomes[2], and contains 20-25,000 genes [7], the simple nematode C elegans has almost as many genes (more than 19,000)[8].
In many species, only a small fraction of the genome encodes protein: only about 1.5% of the human genome consists of protein-coding exons, while over 50% consists of non-coding repetitive sequences.[9] While most of the non-coding elements in human DNA were once considered "junk DNA" with no known function, recent studies suggest a variety of roles for these regions, such as RNA silencing[10] or gene swapping by transposable elements.[11]
Some other vertebrates, including the puffer fish Fugu have very much more compact genomes, and (for multicellular organisms) there seems to be no consistent relationship between the size of the genome and the complexity of the organism [12]. Some non-coding DNA sequences are now known to have a structural role in chromosomes. In particular, telomeres and centromeres contain few genes, but are important for the function and stability of chromosomes.[13][14] An abundant form of non-coding DNA in humans are pseudogenes, which are copies of genes that have been disabled by mutation;[15] these are usually just molecular 'fossils', but they can provide the raw genetic material for new genes.[16]
A recent challenge to the long-standing view that the human genome consists of relatively few genes along with a vast amount of "junk DNA" comes from the ENCyclopedia Of DNA Elements (ENCODE) consortium.[17][18] Their survey of the human genome shows that most of the DNA is transcribed into molecules of RNA. This broad pattern of transcription was unexpected, but whether these transcribed (but not translated) elements have any biological function is not yet clear.[18]
Regulation of gene expression
How genes are regulated — how they are turned on or off — is an important topic in modern biology, and research continually yields surprising insights into how phenotypic traits and biological adaptations are determined.
In the 1950's, investigations of the bacterium Escherichia coli led to the recognition that the region upstream of a transcribed region provides a place for the enzyme RNA polymerase to attach to DNA and start transcribing RNA in the 5' to 3' direction of the nucleic acid chain. The site at which this occurs came to be called the promoter. Other regulatory proteins (such as repressors) influence transcription by binding to a region near to or overlapping the promoter, called the operator. In the early years of modern genetics, emphasis was given to transcriptional regulation as an important and common means of modulating gene expression, but today we realize that there are a wide range of mechanisms by which the expression of mRNA and proteins can be modulated by both external and internal signals in cells.
Physical and chemical properties
DNA is a long chain of repeating units called nucleotides (a nucleotide is a base linked to a sugar and one or more phosphate groups)[19] The DNA chain is 22-26 Å wide (2.2-2.6nm)[20] The nucleotides are very small (just 3.3Å long), but DNA can contain millions of them: the DNA in the largest human chromosome (Chromosome 1) has 220 million base pairs.[21]
In living organisms, DNA does not usually exist as a single molecule, but as a tightly-associated pair of molecules.[22][23] These two long strands are entwined in the shape of a double helix. DNA can thus be thought of as an anti-parallel double helix. The nucleotide repeats contain both the backbone of the molecule, which holds the chain together, and a base, which interacts with the other DNA strand. The double helix is held together by hydrogen bonds between the bases attached to the two strands. If many nucleotides are linked together, as in DNA, the polymer is referred to as a polynucleotide.[24]
The backbone of the DNA strand has alternating phosphate and sugar residues[25]: the sugar is the pentose (five carbon) sugar 2-deoxyribose. The sugar molecules are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms in the sugar rings. Because these bonds are asymmetric, a strand of DNA has a 'direction'. In a double helix, the direction of the nucleotides in one strand is opposite to that in the other strand. This arrangement of DNA strands is called antiparallel. The asymmetric ends of a strand of DNA bases are referred to as the 5' (five prime) and 3' (three prime) ends. One of the major differences between DNA and RNA is the sugar: 2-deoxyribose is replaced by ribose in RNA.[23]
The four bases in DNA are adenine (A), cytosine (C), guanine (G) and thymine (T), and these bases are attached to the sugar/phosphate to form the complete nucleotide. Adenine and guanine are fused five- and six-membered heterocyclic compounds called purines, while cytosine and thymine are six-membered rings called pyrimidines.[24] A fifth pyrimidine base, uracil (U), replaces thymine in RNA. Uracil is normally only found in DNA as a breakdown product of cytosine, but bacterial viruses contain uracil in their DNA.[26] In contrast, following synthesis of certain RNA molecules, many uracils are converted to thymines. This occurs mostly on structural and enzymatic RNAs like tRNAs and ribosomal RNA.[27]
The double helix is a right-handed spiral. As the DNA strands wind around each other, gaps between the two phosphate backbones reveal the sides of the bases inside (see animation). Two of these grooves twist around the surface of the double helix: the major groove is 22 Å wide and the minor groove is 12 Å wide.[28] The edges of the bases are more accessible in the major groove, so proteins like transcription factors that can bind to specific sequences in double-stranded DNA usually make contacts to the sides of the bases exposed in the major groove.[29] For many DNA: protein complexes, up to one-half of the contacts involve the DNA phosphate groups, and thus many DNA binding proteins have more positive amino acids (lysine and arginine) than usual.
Base pairing
Each type of base on one strand of DNA forms a bond with just one type of base on the other strand, called 'complementary base pairing'. Purines form hydrogen bonds to pyrimidines; for example, A bonds only to T, and C bonds only to G. This arrangement of two nucleotides joined together across the double helix is called a base pair. In a double helix, the two strands are also held together by hydrophobic effects and by a variation of pi stacking.[30] Hydrogen bonds can be broken and rejoined quite easily, so the two strands of DNA in a double helix can be pulled apart like a zipper, either by mechanical force or by high temperatures.[31] Becauseof this complementarity, the information in the double-stranded sequence of a DNA helix is duplicated on each strand, and this is vital in DNA replication. The reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in living organisms.[19]
The two types of base pairs form different numbers of hydrogen bonds; AT forms two hydrogen bonds, and GC forms three, so the GC base-pair is stronger than the AT pair. Thus, long DNA helices with a high GC content have strongly interacting strands, while short helices with high AT content have weakly interacting strands.[32] Parts of the DNA double helix that need to separate easily tend to have a high AT content, making the strands easier to pull apart.[33] The strength of this interaction can be measured by finding the temperature required to break the hydrogen bonds (their 'melting temperature'). When all the base pairs in a DNA double helix melt, the strands separate, leaving two single-stranded molecules in solution. Because base-pairing causes changes in the UV-visible spectrum of the bases, DNA melting can be monitored by observing the hypochronicity (or loss thereof) as a function of temperature. These molecules have no single shape, but some conformations are more stable than others.
Sense and antisense
DNA is converted into RNA by RNA polymerases.[34] A DNA sequence is called a "sense" sequence if it is converted into RNA by these enzymes (which only work in the 5' to 3' direction) which is then translated into protein. The sequence on the opposite strand is complementary to the sense sequence and is called the "antisense" sequence. Sense and antisense sequences can co-exist on the same strand of DNA; in both prokaryotes and eukaryotes, antisense sequences are transcribed[35], and antisense RNAs might be involved in regulating gene expression.[36]. (See Micro RNA, RNA interference, sRNA.)
Many DNA sequences in prokaryotes and eukaryotes (and more in plasmids and viruses) have overlapping genes which may both occur in the same direction, on the same strand (parallel) or in opposite directions, on opposite strands (antiparallel).[37][38] In these cases, some DNA sequences encode one protein when read from 5′ to 3′ along one strand, and a different protein when read in the opposite direction (but still from 5′ to 3′) along the other strand. In bacteria, this overlap may be involved in regulating gene transcription,[38] while in viruses, overlapping genes increase the information that can be encoded within the small viral genome.[39] Another way of reducing genome size is seen in some viruses that contain linear or circular single-stranded DNA.[40]
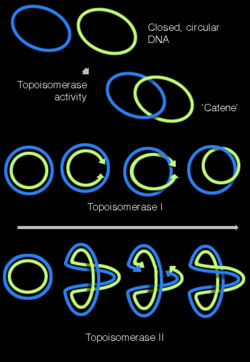
Supercoiling
DNA can be 'twisted' in a process called DNA supercoiling. In its "relaxed" state, a DNA strand usually circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted, the strands become more tightly or more loosely wound.[41] If the DNA is twisted in the direction of the helix (positive supercoiling), and the bases are held more tightly together. If they are twisted in the opposite direction (negative supercoiling) the bases come apart more easily. Most DNA has slight negative supercoiling that is introduced by topoisomerases. These enzymes are also needed to relieve the twisting stresses introduced into DNA strands during processes such as transcription and DNA replication.[42]
Alternative conformations
The conformation of a DNA molecule depends on its sequence, the amount and direction of supercoiling, chemical modifications of the bases, and also solution conditions, such as the concentration of metal ions.[43] Accordingly, DNA can exist in several possible conformations, but only a few of these ("A-DNA", "B-DNA", and "Z-DNA") are thought to occur naturally. Of these, the "B" form is most common. The A form is a wider right-handed spiral, with a shallow and wide minor groove and a narrower and deeper major groove; this form occurs in dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, as well as in enzyme-DNA complexes.[44] Segments of DNA where the bases have been modified by methylation may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form.[45] These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in regulating transcription.[46]
Quadruplex structures
At the ends of the linear chromosomes, specialized regions called telomeres allow the cell to replicate chromosome ends using the enzyme telomerase.[47] Without telomeres, a chromosome would become shorter each time it was replicated. These specialized 'caps' also help to protect the DNA ends from exonucleases and stop the DNA repair systems in the cell from treating them as damage to be corrected. In human cells, telomeres are usually lengths of single-stranded DNA that contain several thousand repeats of a TTAGGG sequence.[13] These sequences may stabilize chromosome ends by forming unusual quadruplex structures. Here, four guanine bases form a flat plate, throughhydrogen bonding, and these plates then stack on top of each other to form a stable quadruplex.[48] Other structures can also be formed, and the central set of four bases can come from either one folded strand, or several different parallel strands, each contributing one base to the central structure.
Telomeres also form large loops called 'telomere loops', or 'T-loops'. Here, the single-stranded DNA curls around in a circle stabilized by telomere-binding proteins.[49]
Chemical modifications
DNA methylation
The expression of genes is influenced by the chromatin structure of a chromosome, and regions of heterochromatin (with little or no gene expression) correlate with the methylation of cytosine.[50] These structural changes to the DNA are one type of epigenetic change that can alter chromatin structure, and they are inheritable. Epigenetics refers to features of organisms that are stable over successive rounds of cell division but which do not involve changes in the underlying DNA sequence[51]. Epigenetic changes are important in cellular differentiation, allowing cells to maintain different characteristics despite containing the same genomic material. Some epigenetic features can be inherited from one generation to the next.[52]
The level of methylation varies between organisms; the nematode C. elegans lacks any cytosine methylation, while up to 1% of the DNA of vertebrates contains 5-methylcytosine.[53] Despite the biological importance of 5-methylcytosine, it is susceptible to spontaneous deamination, and methylated cytosines are therefore mutation 'hotspots'.[54] Other base modifications include adenine methylation in bacteria and the glycosylation of uracil to produce the "J-base" in kinetoplastids.[55][56]
Mutations
DNA can be damaged by many different agents. Mutagens are agents which can produce genetic mutations - these are alterations of one DNA base to another base. Examples of mutagens include oxidizing agents, alkylating agents and high-energy electromagnetic radiation such as ultraviolet light and x-rays. Lesions of DNA, in which residues are changed to a structure that is not a normal feature of DNA, are different from mutations. However, damaged DNA can give rise to mutations, and indeed, some DNA repair processes are error prone and thus themselves generate mutations.
Ultraviolet radiation damages DNA mostly by producing thymine dimers.[57] Oxidants such as free radicals or hydrogen peroxide can cause several forms of damage, including base modifications (particularly of guanosine) as well as double-strand breaks.[58] It has been estimated that, in each human cell, about 500 bases suffer oxidative damage every day.[59][60] Of these lesions, the most damaging are double-strand breaks, as they can produce point mutations, insertions and deletions from the DNA sequence, as well as chromosomal translocations.[61]
Many mutagens intercalate into the space between two adjacent base pairs. These are mostly polycyclic, aromatic, and planar molecules, and include ethidium, proflavin, daunomycin, doxorubicin and thalidomide. For an intercalator to fit between base pairs, the bases must separate, distorting the DNA strand by unwinding of the double helix. These structural modifications inhibit transcription and replication processes, causing both toxicity and mutations. As a result, DNA intercalators are often carcinogens. Nevertheless, because they can inhibit DNA transcription and replication, they are also used in chemotherapy to inhibit DNA-replication in rapidly-growing cancer cells.[62]
Interactions with proteins
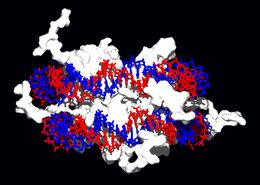
All of the functions of DNA depend on its interactions with proteins. some of these interactions are non-specific, others are specific in that the protein can only bind to a particular DNA sequence. Some enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
DNA-binding proteins
|
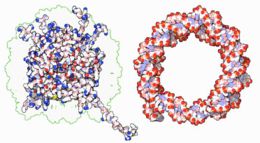
|
Within chromosomes, DNA is held in complexes between DNA and structural proteins. These proteins organize the DNA into a compact structure called chromatin. In eukaryotes, this structure involves DNA binding to small basic proteins called histones, while in prokaryotes many types of proteins are involved.[63] The histones form a disk-shaped complex called a nucleosome which has two complete turns of double-stranded DNA wrapped around it. These interactions are formed through basic residues in the histones making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are largely independent of the base sequence.[64] Chemical modifications of these basic amino acid residues include methylation, phosphorylation and acetylation.[65] These changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to transcription factors and changing the rate of transcription.[66] Other non-specific DNA-binding proteins found in chromatin include the high-mobility group proteins, which bind preferentially to bent or distorted DNA.[67] These proteins are important in bending arrays of nucleosomes and arranging them into more complex chromatin structures.[68]
Another group of DNA-binding proteins bind single-stranded DNA. In humans, replication protein A is the best-characterised member of this family, and is essential for most processes where the double helix is separated (including DNA replication, recombination and DNA repair).[69] These proteins seem to stabilize single-stranded DNA and protect it from forming stem loops or being degraded by nucleases.
Other proteins bind to particular DNA sequences. The most intensively studied of these are the transcription factors. Each of these proteins binds to a particular set of DNA sequences and thereby activates or inhibits the transcription of genes with these sequences close to their promoters. Transcription factors do this in two ways. Some can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription.[70] Other transcription factors can bind enzymes that modify the histones at the promoter, this will change the accessibility of the DNA template to the polymerase.[71]
These DNA targets can occur throughout an organism's genome, so changes in the activity of one type of transcription factor in a given cell can affect the expression of many genes in that cell.[72] Consequently, these proteins are often the targets of the signal transduction processes that mediate responses to environmental changes or cellular differentiation and development. The specificity of transcription factor interactions with DNA arises because the proteins make multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these interactions occur in the major groove, where the bases are most accessible.[29]
DNA-modifying enzymes
Nucleases and ligases
Nucleases cut DNA strands by catalyzing the hydrolysis of the phosphodiester bonds (nucleases that hydrolyse nucleotides from the ends of DNA strands are called exonucleases, while endonucleases cut within strands). The most frequently-used nucleases in molecular biology are the restriction endonucleases, which cut DNA at specific sequences. For instance, the EcoRV enzyme recognizes the 6-base sequence 5′-GAT|ATC-3′ and makes a cut at the vertical line. In nature, these enzymes protect bacteria against phage infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the restriction modification system.[73] In technology, these sequence-specific nucleases are used in molecular cloning and DNA fingerprinting.
DNA ligases can rejoin cut or broken DNA strands, using the energy from either adenosine triphosphate or nicotinamide adenine dinucleotide. Ligases are particularly important in lagging strand DNA replication, as they join together the short segments of DNA produced at the replication fork into a complete copy of the DNA template. They are also used in DNA repair and genetic recombination.[74]
Topoisomerases and helicases
Topoisomerases have both nuclease and ligase activity, and they can change the amount of supercoiling in DNA. Some work by cutting the DNA helix and allowing one section to rotate; the enzyme then seals the DNA break.[75] Others can cut one DNA helix and then pass a second strand of DNA through this break, before rejoining the helix.[76] Topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.[42]
Helicases are a type of 'molecular motor'; they use the chemical energy in nucleoside triphosphates (predominantly ATP) to break hydrogen bonds between bases and unwind the DNA double helix into single strands.[77] These enzymes are essential for most processes where enzymes need to access the DNA bases.
Polymerases
Polymerases synthesise polynucleotides from nucleoside triphosphates. They add nucleotides onto the 3′ hydroxyl group of the previous nucleotide in the DNA strand, so all polymerases work in a 5′ to 3′ direction.[34] In the active site of these enzymes, the nucleoside triphosphate substrate base-pairs to a single-stranded polynucleotide template: this allows polymerases to synthesise the complementary strand of this template accurately.
DNA polymerases copy DNA sequences. Accuracy is very important for this, so many polymerases have a proofreading activity: they can recognizes the occasional mistakes that can occur during synthesis. These enzymes detect the lack of base pairing between mismatched nucleotides, and if a mismatch is detected, a 3′ to 5′ exonuclease activity is activated and the incorrect base is removed.[78] In most organisms, DNA polymerases function in a large complex called the replisome.
RNA-dependent DNA polymerases copy the sequence of an RNA strand into DNA. One example is reverse transcriptase, which is a viral enzyme involved in the infection of cells by retroviruses; another example is telomerase, which is required for the replication of telomeres.[79][47] Telomerase is an unusual polymerase because it contains its own RNA template as part of its structure.[80]
Transcription is carried out by a RNA polymerase that copies a DNA sequence into RNA. The enzyme binds to a promoter and separates the DNA strands. It then copies the gene sequence into a mRNA transcript until it reaches a region of DNA called the terminator, where it halts and detaches from the DNA. RNA polymerase II, which transcribes most of the genes in the human genome, operates as part of a large protein complex with multiple regulatory and accessory subunits.[81]
Genetic recombination
A DNA helix does not usually interact with other segments of DNA, and in human cells the different chromosomes even occupy different regions of the nucleus (called "chromosome territories").[82] This physical separation of chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is when they recombine. Recombination is when two DNA helices break, swap a section and then rejoin. In eukaryotes, this usually occurs during meiosis, when two chromatids are paired together in the center of the cell. This allows chromosomes to exchange genetic information and produces new combinations of genes.[83] Genetic recombination can also be involved in DNA repair.[84] The most common form of recombination is homologous recombination, where the two chromosomes involved share very similar sequences. However, recombination can also damage cells, by producing chromosomal translocations and genetic abnormalities. Recombination reactions are catalyzed by recombinases,[85] which have a DNA-dependent ATPase activity. The recombinase makes a 'nick' in one strand of a DNA double helix, allowing the nicked strand to separate from its complementary strand and anneal to one strand of the double helix on the opposite chromatid. A second nick allows the strand in the second chromatid to separate and anneal to the strand in the first helix, forming a cross-strand exchange (also called a Holliday junction). The Holliday junction is a tetrahedral junction structure which can be moved along the pair of chromosomes, swapping one strand for another. The recombination is then halted by cleavage of the junction and re-ligation of the released DNA.[86]
DNA and molecular evolution
As well as being susceptible to largely random mutations (that usually affect just a single base), some regions of DNA are specialised to undergo dramatic, rapid, non-random rearrangements, or to undergo more subtle changes at a high frequency so that the expression of a gene is dramatically altered. Such rearrangements include various versions of site-specific recombination. This depends on enzymes that recognise particular sites on DNA and create novel structures such as insertions, deletions and inversions. For instance, DNA regions in the small genome of the bacterial virus P1 can invert, enabling different versions of tail fibers to be expressed in different viruses. Similarly, site-specific recombinase enzymes are responsible for mating type variation in yeast, and for flagellum type (phase) changes in the bacterium Salmonella enterica Typhimurium.
More subtle mutations can occur in micro-satellite repeats (also called homopolymeric tracts of DNA). An example of such a structure are short DNA intervals where the same base is tandemly repeated, as in 5'-gcAAAAAAAAAAAttg-3', or a dinucleotide or even a triplet as in 5'- ATGATGATGATGATGATGATG-3'. DNA polymerase III is prone to make 'stuttering errors' at such repeats. As a consequence, changes in repeat number occur quite often during cell replication, and when they appear at a position where the spacing of nucleotide residues is critical for gene function, they can cause changes to the phenotype.
The stomach ulcer bacterium Helicobacter pylori is a good example of how homopolymeric tracts can enable quasi-directed evolution of particular genes. H. pylori has 46 genes that contain homopolymeric runs of nucleotides or dinucleotide repeats that are prone to frequent length changes as a consequence of stuttering errors during replication. These changes can lead to frequent reversible inactivation of these genes, or to changed gene transcription if the repeat is located in a regulatory sequence. This generates highly diverse populations of H. pylori in an individual human host, and this diversity helps the bacterium evade the immune system.[87]
Triplet repeats behave similarly, but are particularly suited to evolution of proteins with differing characteristics. In the clock-like period gene of the fruit fly, triplet repeats 'fine tune' an insect's biological clock in response to changes in environmental temperatures. Triplet repeats are widely distributed in genomes, and their high frequency of mutation is responsible for several genetically determined disorders in humans.[88]
Uses in technology
Forensics
Forensic scientists can use DNA in blood, semen, skin, saliva or hair to match samples collected at a crime scene to samples taken from possible suspects. This process is called genetic fingerprinting or more formally, DNA profiling. In DNA profiling, the lengths of variable sections of repetitive DNA (such as short tandem repeats and minisatellites) are compared between people. This is usually very reliable for identifying the source of a sample,[89] but identification can be complicated if the samples that are collected include DNA from several people.[90] DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys,[91] and first used in forensic science to convict Colin Pitchfork (and to clear the prime suspect) in the 1988 Enderby murders case.[92] People convicted of certain types of crimes may be required to provide a sample of DNA for a database. This has helped investigators solve old cases where only a DNA sample was obtained from the scene. DNA profiling can also be used to identify victims of mass casualty incidents.[93]
Bioinformatics
One branch of bioinformatics involves the analysis of DNA sequence data; DNA from hundreds of different organisms has now been partially sequenced, and this information is stored in massive databases. The development of techniques to store and search DNA sequences has led to many applications in computer science.[94] String-searching or 'matching' algorithms, which identify a given sequence of letters inside a larger sequence of letters, are used to search for specific sequences of nucleotides. The related problem of sequence alignment aims to identify homologous sequences; these are sequences that are very similar but not identical. When two different genes in an organism have very similar sequences, this is evidence that, at some stage in evolution, a single gene was duplicated, and the sequences subsequently diverged (under different selection pressures) by incorporating different mutations. Identifying such holologies can give valuable clues about the likely function of novel genes. Similarly, identifying homologies between genes in different organisms can be used to reconstruct the evolutionary relationships between organisms.[95] Data sets representing entire genomes' worth of DNA sequences, such as those produced by the Human Genome Project, are difficult to use without annotations, which label the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have patterns that are characteristic of protein- or RNA-coding genes can be identified by gene finding algorithms, allowing researchers to predict the presence of particular gene products in an organism.[96]
Molecular cloning
The small chromosomes of bacteria have proved extraordinarily useful for analysing genetic mechanisms and genome structure in many organisms, including humans. This utility arose because of several important discoveries made by microbiologists in the 1950-70's that provided new laboratory tools for directly manipulating genes. The principal laboratory tools and techniques discovered in this period were:
- Compact circular plasmid molecules such as ColE1 that were relatively easy to extract and purify from bacterial cells, and when inserted back inside living bacterial cells could serve as genetically stable carriers (vectors) of novel DNA fragments. These vectors were propagated in bacterial cell lines termed "clones". Since each plasmid vector in these clones carried a single DNA molecular fragment, these could be considered to be 'molecular clones'.
- Convenient methods for extracting plasmid DNA from cells, physical analysis of this DNA, and reinsertion of circular plasmid DNA back inside living cells. The main method for DNA re-insertion is termed DNA transformation - that is, direct uptake of naked DNA molecules by cells. One technique that assisted DNA transformation was the use of selectable genetic markers, and plasmid-borne bacterial antibiotic resistance provided such markers in the form of traits like ampicillin resistance and tetracycline resistance.
- Restriction endonucleases such as the enzymes EcoRI or HindIII, were found to be useful for specifically digesting DNA at particular sites and creating novel combinations of DNA fragments in the laboratory.
- Restriction enzymes enabled re-annealing different DNA fragments, such as circular plasmid DNA which had been linearised at a single EcoRI restriction endonuclease target site together with another fragment, say an EcoRI generated fragment of a human chromosome. By sealing together two such fragments with the enzyme DNA ligase, novel hybrid-plasmid molecules could be created in which "foreign" DNA inserts were carried in a chimeric or hybrid plasmid, often called a recombinant DNA molecule.
After the mid-1970's, re-insertion of recombinant DNA plasmids into living bacterial cells, such as those of Escherichia coli opened up many new approaches in biotechnology. These made it possible to manufacture, for example, human hormones such as insulin in microbial cell-based protein factories. When combined with other genetic techniques, the field of "recombinant DNA technology" opened up the possibility of decoding the gene sequence of whole genomes. This latter area is now usually referred to as genomics, and includes the Human Genome Project.
References
- ↑ Kornberg, Arthur. "DNA Replication". W.H.Freeman and Co. (1980) p13
- ↑ 2.0 2.1 Venter J et al. (2001). "The sequence of the human genome". Science 291: 1304–51. PMID 11181995.
- ↑ Human Genome Project Information [1]
- ↑ Albà M (2001). "Replicative DNA polymerases". Genome Biol 2: REVIEWS3002. PMID 11178285.
- ↑ Pollard KS et al. (2006) Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2(10):e168 PMID 17040131
- ↑ Thanbichler M et al. (2005). "The bacterial nucleoid: a highly organized and dynamic structure". J Cell Biochem 96: 506–21. PMID 15988757.
- ↑ Human Genome Project Information [2]
- ↑ The C. elegans Sequencing Consortium (1998)Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology. Science 282: 2012-8 [3]
- ↑ Wolfsberg T et al. (2001). "Guide to the draft human genome". Nature 409: 824-6. PMID 11236998.
- ↑ Volpe, TA "et al." (2002). "Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi". Science 297: 1833-1837.
- ↑ name=Biemont>Biemont, C and Vieira, C (2006). "Genetics: Junk DNA as an evolutionary force". Nature 443: 521-524.
- ↑ See Sydney Brennar's Nobel lecture (2002) [4]
- ↑ 13.0 13.1 Wright W et al. (1997). "Normal human chromosomes have long, G-rich telomeric overhangs at one end". Genes Dev 11: 2801-9. PMID 9353250.
- ↑ Pidoux A, Allshire R (2005). "The role of heterochromatin in centromere function". Philos Trans R Soc Lond B 360: 569-79. PMID 15905142.
- ↑ Harrison P et al. (2002). "Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22". Genome Res 12: 272-80. PMID 11827946.
- ↑ Harrison P, Gerstein M (2002). "Studying genomes through the aeons: protein families, pseudogenes and proteome evolution". J Mol Biol 318: 1155-74. PMID 12083509.
- ↑ Weinstock GM (2007). "ENCODE: More genomic empowerment". Genome Research 17: 667-668. PMID 17567987.
- ↑ 18.0 18.1 ENCODE Project Consortium (2007). "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project". Nature 447: 799-816. PMID 17571346.
- ↑ 19.0 19.1 Alberts, Bruce; Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walters (2002). Molecular Biology of the Cell; Fourth Edition. New York and London: Garland Science. ISBN 0-8153-3218-1.
- ↑ Mandelkern M et al. (1981). "The dimensions of DNA in solution". J Mol Biol 152: 153-61. PMID 7338906.
- ↑ Gregory S et al. (2006). "The DNA sequence and biological annotation of human chromosome 1". Nature 441: 315-21. PMID 16710414.
- ↑ Watson J, Crick F (1953). "Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid". Nature 171: 737-8. PMID 13054692.
- ↑ 23.0 23.1 Berg J et al. (2002) Biochemistry. WH Freeman and Co. ISBN 0-7167-4955-6
- ↑ 24.0 24.1 Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents IUPAC-IUB Commission on Biochemical Nomenclature (CBN) Accessed 03 Jan 2006
- ↑ Ghosh A, Bansal M (2003). "A glossary of DNA structures from A to Z". Acta Crystallogr D Biol Crystallogr 59: 620-6. PMID 12657780.
- ↑ Takahashi I, Marmur J (1963). "Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis". Nature 197: 794-5. PMID 13980287.
- ↑ Agris P (2004). "Decoding the genome: a modified view". Nucleic Acids Res 32: 223 – 38. PMID 14715921.
- ↑ Wing R et al. (1980). "Crystal structure analysis of a complete turn of B-DNA". Nature 287: 755 – 8. PMID 7432492.
- ↑ 29.0 29.1 Pabo C, Sauer R. "Protein-DNA recognition". Ann Rev Biochem 53: 293-321. PMID 6236744.
- ↑ Ponnuswamy P, Gromiha M (1994). "On the conformational stability of oligonucleotide duplexes and tRNA molecules". J Theor Biol 169: 419-32. PMID 7526075.
- ↑ Clausen-Schaumann H et al. (2000). "Mechanical stability of single DNA molecules". Biophys J 78: 1997-2007. PMID 10733978.
- ↑ Chalikian T et al. (1999). "A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques". Proc Natl Acad Sci USA 96: 7853-8. PMID 10393911.
- ↑ deHaseth P, Helmann J (1995). "Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA". Mol Microbiol 16: 817-24. PMID 7476180.
- ↑ 34.0 34.1 Joyce C, Steitz T (1995). "Polymerase structures and function: variations on a theme?". J Bacteriol 177: 6321-9. PMID 7592405.
Cite error: Invalid
<ref>tag; name "Joyce" defined multiple times with different content - ↑ Hüttenhofer A et al. (2005). "Non-coding RNAs: hope or hype?". Trends Genet 21: 289-97. PMID 15851066.
- ↑ Munroe S (2004). "Diversity of antisense regulation in eukaryotes: multiple mechanisms, emerging patterns". J Cell Biochem 93: 664-71. PMID 15389973.
- ↑ Makalowska I et al. (2005). "Overlapping genes in vertebrate genomes". Comput Biol Chem 29: 1–12. PMID 15680581.
- ↑ 38.0 38.1 Johnson Z, Chisholm S (2004). "Properties of overlapping genes are conserved across microbial genomes". Genome Res 14: 2268–72. PMID 15520290.
- ↑ Lamb R, Horvath C (1991). "Diversity of coding strategies in influenza viruses". Trends Genet 7: 261–6. PMID 1771674.
- ↑ Davies J, Stanley J (1989). "Geminivirus genes and vectors". Trends Genet 5: 77–81. PMID 2660364.
- ↑ Benham C, Mielke S. "DNA mechanics". Ann Rev Biomed Eng 7: 21–53. PMID 16004565.
- ↑ 42.0 42.1 Wang J (2002). "Cellular roles of DNA topoisomerases: a molecular perspective". Nat Rev Mol Cell Biol 3: 430–40. PMID 12042765.
- ↑ Basu H et al. (1988). "Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: experimental and theoretical studies". J Biomol Struct Dyn 6: 299-309. PMID 2482766.
- ↑ Lu XJ et al. (2000). "A-form conformational motifs in ligand-bound DNA structures". J Mol Biol 300: 819-40. PMID 10891271.
- ↑ Rothenburg S et al.. "DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles". Immunol Rev 184: 286–98. PMID 12086319.
- ↑ Oh D et al. (2002). "Z-DNA-binding proteins can act as potent effectors of gene expression in vivo". Proc Natl Acad Sci USA 99: 16666-71. PMID 12486233.
- ↑ 47.0 47.1 Greider C, Blackburn E (1985). "Identification of a specific telomere terminal transferase activity in Tetrahymena extracts". Cell 43: 405-13. PMID 3907856.
- ↑ Burge S et al. (2006). "Quadruplex DNA: sequence, topology and structure". Nucleic Acids Res 34: 5402-15. PMID 17012276.
- ↑ Griffith J et al. (1999). "Mammalian telomeres end in a large duplex loop". Cell 97: 503-14. PMID 10338214.
- ↑ For example, cytosine methylation, to produce 5-methylcytosine, is important for X-chromosome inactivation. Klose R, Bird A (2006). "Genomic DNA methylation: the mark and its mediators". Trends Biochem Sci 31: 89–97. PMID 16403636.
- ↑ Adrian Bird (2007). "Perceptions of epigenetics". Nature 447: 396-398. PMID 17522671
- ↑ V.L. Chandler (2007). "Paramutation: From Maize to Mice". Cell 128: 641-645.
- ↑ Bird A (2002). "DNA methylation patterns and epigenetic memory". Genes Dev 16: 6–21. PMID 11782440.
- ↑ Walsh C, Xu G. "Cytosine methylation and DNA repair". Curr Top Microbiol Immunol 301: 283–315. PMID 16570853.
- ↑ Ratel D et al. (2006). "N6-methyladenine: the other methylated base of DNA". Bioessays 28: 309–15. PMID 16479578.
- ↑ Gommers-Ampt J et al. (1993). "beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei". Cell 75: 1129–36. PMID 8261512.
- ↑ Douki T et al. (2003). "Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar ultraviolet radiation". Biochemistry 42: 9221-6. PMID 12885257.
- ↑ Cadet J et al. (1999). "Hydroxyl radicals and DNA base damage". Mutat Res 424 (1-2): 9-21. PMID 10064846.
- ↑ Shigenaga M et al. (1989). "Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage". Proc Natl Acad Sci USA 86: 9697-701. PMID 2602371.
- ↑ Cathcart R et al. (1984). "Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage". Proc Natl Acad Sci USA 81: 5633-7. PMID 6592579.
- ↑ Valerie K, Povirk L (2003). "Regulation and mechanisms of mammalian double-strand break repair". Oncogene 22: 5792-812. PMID 12947387.
- ↑ Braña M et al. (2001). "Intercalators as anticancer drugs". Curr Pharm Des 7: 1745-80. PMID 11562309.
- ↑ Sandman K et al. (1998). "Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome". Cell Mol Life Sci 54: 1350-64. PMID 9893710.
- ↑ Luger K et al. (1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature 389: 251-60. PMID 9305837.
- ↑ Jenuwein T, Allis C (2001). "Translating the histone code". Science 293: 1074-80. PMID 11498575.
- ↑ Ito T. "Nucleosome assembly and remodelling". Curr Top Microbiol Immunol 274: 1-22. PMID 12596902.
- ↑ Thomas J (2001). "HMG1 and 2: architectural DNA-binding proteins". Biochem Soc Trans 29: 395-401. PMID 11497996.
- ↑ Grosschedl R et al. (1994). "HMG domain proteins: architectural elements in the assembly of nucleoprotein structures". Trends Genet 10: 94-100. PMID 8178371.
- ↑ Iftode C et al. (1999). "Replication protein A (RPA): the eukaryotic SSB". Crit Rev Biochem Mol Biol 34: 141-80. PMID 10473346.
- ↑ Myers L, Kornberg R. "Mediator of transcriptional regulation". Ann Rev Biochem 69: 729-49. PMID 10966474.
- ↑ Spiegelman B, Heinrich R (2004). "Biological control through regulated transcriptional coactivators". Cell 119: 157-67. PMID 15479634.
- ↑ Li Z et al. (2003). "A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells". Proc Natl Acad Sci USA 100: 8164-9. PMID 12808131.
- ↑ Bickle T, Krüger D (1993). "Biology of DNA restriction". Microbiol Rev 57: 434–50. PMID 8336674.
- ↑ Doherty A, Suh S (2000). "Structural and mechanistic conservation in DNA ligases.". Nucleic Acids Res 28: 4051–8. PMID 11058099.
- ↑ Champoux J. "DNA topoisomerases: structure, function, and mechanism". Ann Rev Biochem 70: 369–413. PMID 11395412.
- ↑ Schoeffler A, Berger J (2005). "Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism". Biochem Soc Trans 33: 1465–70. PMID 16246147.
- ↑ Tuteja N, Tuteja R (2004). "Unraveling DNA helicases. Motif, structure, mechanism and function". Eur J Biochem 271: 1849–63. PMID 15128295.
- ↑ Hubscher U et al.. "Eukaryotic DNA polymerases". Annu Rev Biochem 71: 133–63. PMID 12045093.
- ↑ Tarrago-Litvak L et al. (1994). "The reverse transcriptase of HIV-1: from enzymology to therapeutic intervention". FASEB J 8: 497–503. PMID 7514143.
- ↑ Nugent C, Lundblad V (1998). "The telomerase reverse transcriptase: components and regulation". Genes Dev 12 (8): 1073–85. PMID 9553037.
- ↑ Martinez E (2002). "Multi-protein complexes in eukaryotic gene transcription". Plant Mol Biol 50: 925–47. PMID 12516863.
- ↑ Cremer T, Cremer C (2001). "Chromosome territories, nuclear architecture and gene regulation in mammalian cells". Nat Rev Genet 2: 292-301. PMID 11283701.
- ↑ Pál C et al. (2006). "An integrated view of protein evolution". Nat Rev Genet 7: 337-48. PMID 16619049.
- ↑ O'Driscoll M, Jeggo P (2006). "The role of double-strand break repair - insights from human genetics". Nat Rev Genet 7: 45-54. PMID 16369571.
- ↑ Sung P et al. (2003). "Rad51 recombinase and recombination mediators". J Biol Chem 278: 42729-32. PMID 12912992.
- ↑ Dickman M et al. (2002). "The RuvABC resolvasome". Eur J Biochem 269: 5492-501. PMID 12423347.
- ↑ Suerbaum S, Josenhans C (2007) Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol 5:441-52 PMID 17505524
- ↑ Christopher Wills discusses the evolutionary significance of these concepts in The Runaway Brain: The Evolution of Human Uniqueness ISBN 0-00-654672-2 (1995)
- ↑ Collins A, Morton N (1994). "Likelihood ratios for DNA identification". Proc Natl Acad Sci USA 91: 6007-11. PMID 8016106.
- ↑ Weir B et al. (1997). "Interpreting DNA mixtures". J Forensic Sci 42: 213-22. PMID 9068179.
- ↑ Jeffreys A et al. (1985). "Individual-specific 'fingerprints' of human DNA.". Nature 316: 76-9. PMID 2989708.
- ↑ Colin Pitchfork - first murder conviction on DNA evidence also clears the prime suspect Forensic Science Service Accessed 23 Dec 2006
- ↑ DNA Identification in Mass Fatality Incidents. National Institute of Justice (September 2006).
- ↑ Baldi, Pierre. Brunak, Soren. Bioinformatics: The Machine Learning Approach MIT Press (2001) ISBN 978-0-262-02506-5
- ↑ Sjölander K (2004). "Phylogenomic inference of protein molecular function: advances and challenges". Bioinformatics 20: 170-9. PMID 14734307.
- ↑ Mount DM (2004). Bioinformatics: Sequence and Genome Analysis, 2. Cold Spring Harbor Laboratory Press. ISBN 0879697121.