Salmonella enterica: Difference between revisions
imported>Debra B. Sherman |
imported>Debra B. Sherman |
||
| Line 41: | Line 41: | ||
==References== | ==References== | ||
[Sample reference] [http://ijs.sgmjournals.org/cgi/reprint/50/2/489 Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. "''Palaeococcus ferrophilus'' gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney". ''International Journal of Systematic and Evolutionary Microbiology''. 2000. Volume 50. p. 489-500.] | [Sample reference] [http://ijs.sgmjournals.org/cgi/reprint/50/2/489 Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. "''Palaeococcus ferrophilus'' gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney". ''International Journal of Systematic and Evolutionary Microbiology''. 2000. Volume 50. p. 489-500.] | ||
[http://jmm.sgmjournals.org/cgi/reprint/56/2/277?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=1&title=typhoid&andorexacttitle=and&andorexacttitleabs=and&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&fdate=1/1/2003&resourcetype=HWCIT] | |||
Revision as of 14:04, 31 March 2008
Articles that lack this notice, including many Eduzendium ones, welcome your collaboration! |
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae
Species
Salmonella enterica
Description and significance
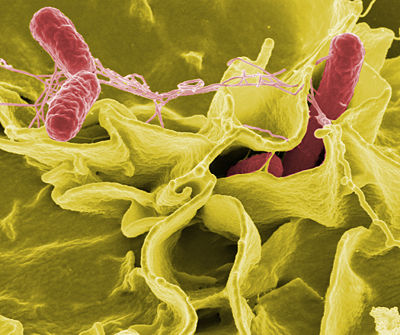
The Salmonella bacteria were first discovered by an American scientist, Dr. Daniel E. Salmon in 1884. Dr. Salmon isolated the bacteria from the intestines of a pig and called it Salmonella choleraesui. The genus Salmonella is divided into two species, S. enterica and S. bongori. Salmonella enterica are rod shaped gram-negative bacteria. Salmonella are facultative anaerobic bacteria . They are very commonly found in raw meat, chicken, and egg shells. Another one of its' habitats is in contaminated water.Once it enters the host, it resides in the intestinal tract of the human or animal. Research done on the genomic sequencing of S. enterica can aid in the effectiveness of medications and vaccinations in treating disease. It is usually isolated on a selective medium such as Maconkey agar, XLD agar, XLT agar, or DCA agar. The three main serovars of Salmonella enterica are Typhi, Typhimurium, and Enteritidis. Their DNA are anywhere between 95% and 99% similar. Ways that researchers have used to differentiate between the different serovars is by looking at their phage specifity. S. typhi is the serovar responsible for typhoid fever, a lethal disease. This disease is usually found in poor, under developed areas. The most common symtoms are fever, vomiting and possible death. S. typhi is found in contaminated water. A human can become infected by drinking the contaminated water or even by washing other foods with the water. S. typhimurium used to be the most common cause of food poisining. It has many of the similar symptoms as S. typhi although not usually fatal. Humans with a weak immune system may require antiobiotics as treatment. S. enteritidishas recently become the most common cause of food poisoning and gives rise to the same symtoms as S. typhimurium. Additionally, it infects and spreads through flocks of chickens. This causes the Salmonella infection to affect the human intestines once the chicken has been ingested.
Genome structure
The genome of S. typhi CT18 is made up on a large circular shaped chromosome and two plasmid referred to as pHCM1 and pHCM2. The chromosome is 4.8Mb and the plasmids are 218 kb and 106 kb, respectively. These plasmids have different drug resistant bands. S. typhimurium has one chromosome that is also 4.8Mb in length, but only one plasmid, pSLT, that is smaller than the plasmids of S. typhi. S. typhi TY2 contains one chromosome that is 4.7Mb long, but lacks plasmids with drug resistant bands. This strain was used to develope the vaccine for infections caused by S. typhi. Psuedogenes are molecules similar to genes, except that they lost their ability to code for proteins. Since they cannot no longer code for or express proteins, they are considered nonfuntional genes. S. typhi CT18 contains 204 pseudogene. Nine of these pseudogenes are genes found in S. typhi TY2. TY2 and CT18 have 195 pseudogenes in common and TY2 has 11 different ones.
Cell structure and metabolism
S. enterica is a motile, rod shaped bacteria that contain peritrichous flagella and produce hydrogen sulfide. Importany molecules that they produce once inside their hosts, are specific proteins that causes the cells of the intestinal walls that it is invading to become disorganized. The disorganization of the cells caused by the production of these proteins causes the bacteria to become engulfed. The proteins that are produced by the bacteria staple together the hosts' actin molecules so that it will fold around the bacteria. This protein is referred to as Salmonella invasion protein A (SipA). New research done at the University of Georgia has found that Salmonella use hydrogen as an energy source. It was originally thought that hydrogen was eliminated from the body as a waste product. New reaserch has discovred that hydrogen is not a waste product, rather it remains in the body and can used as evergy by invading bacteria. When researchers sequenced the genome of Salmonella bacteria they found that they contained three membrane associated enzymes that helped break down hygrogen.
Ecology
Researchers have discovered certain mechanisms that Salmonella bacteria use in order to allow them to survive in harsh and hostile environments. They possess a regulatory system that consists of two parts known as PmrA and PmrB. They use these systems to detect certain stimuli in the environment and then respond to them by either repressing or activating specific gene expression. Researchers additionally discovered that the PmrA/PmrB complex is activated by the presence of iron. This is very important and beneficial for the survival or the bacteria because this complex offers them a protective response. High iron levels that trigger the activation of the PmrA/PmrB complex can be found in the soil and water. High levels of iron can also be found in the intestinal tracts of various animals and humans. The activation of this complex by iron helps them protect themselves against the antiobiotic polymyxin. Nonetheless, S. enterica can be found in many eukaryotic organsims including humans, animals, birds, and even reptiles. It is excreted in feces and transferred to to other organisms or environmental habitats by contaminated water or soil. S. enterica can survive in environments with a pH value of 4 to 8 and a temperature value between 8 and 45 degrees celcius. They are found to be very resistant to certain food preserving methods like drying, salting and smoking. However, they are not resistant, and in fact very sensitive to certain radiations. This is why they love living in damp, wet soil because it protects them from the sun.
Pathology
Different species of Salmonella can cause a wide range of diseases from gastrointestinal problems to typhoid fever. Usually the strain can be carried in a large range hosts including humans, animals, rodents, and birds. S. typhi, a serovar of S. enterica, will only infect humans. The typhoid fever that it causes kills 500,000 people per year. Other serovars like S. typhimurium infect humans, as well as many other mammalian species. The bacteria enters the host by disturbing the membrane. Once inside, it harms the host by causing the levels of intracellular free calcium to increase and disorganize the cytoplasm of the cell. From the intestines, it is trasnfered to the liver or the spleen where it continues to grow. Then, it either goes back into the hosts's intestines, or is excreted in the organisms' feces. It can be spread from the feces by contaminated water, soil, or poor sanitary conditons. Symptoms usually include diarrhea, fever, and abdominal cramps. Strains that are associated with food-borne infections are usually not treated with antibiotics, but will resolve themselves over time. The typhoidal strains usually do require treatment by antibiotics, and possibly hospitilization. Some of the antibiotics have been used in the beef and poultry industries. This has caused for a strain of bacteria to be resistant to the antibiotics. Suggested ways to prevent infections from Salmonella bacteria include making sure all foods are completely cooked, washing all kitchen surfaces, dishes, and hands thoroughly, refrigerating foods immediatly. A most famouse case of Salmonella is one by Mary Mallon. She is the first known American to be a healthy carried of Typhoid fever. People acquire Typhoid fever after eating or drinking water or food that has been touched by carriers of the disease. A carrier is one who has had Typhoid fever in the past and survived, but yet has the typhoid bacteria in their system without any symtoms. Since the bacteria is still within then, carriers will still excrete the bacteria in their feces, which will then spread and infect other organisms. Mary Mallon, a carrier of the disease, was a cook in New York City in the early 1900's. She infected 24 people with typhoid fever. She then switched jobs and infected and killed 11 more people. She swicthed jobs multiple times, each time infecting people with typhoid fever. A total of 47 people were infected by Mary Mallon and three of them died. Mary denied that she was the cause of the infection, insisting that she was never sick with typhoid fever. Typhoid Mary has become a term used by many to descrbe a carrier or a disease that causes harm to others around them and yet refuses to take responsibiiy. In addition, this one case shows how fast the Salmonella bacteria can spread from one person if proper sanitation methods are not kept.
Application to Biotechnology
Researchers have discovered that Salmonella bacteria can be used to destroy cancerous tumos that cannot be cure by chemotherapy. The use a mutant strain of S. typhimurium so that it will be harmless to the host. Bacteria live very comfortably in tumors due its environment which is very low in oxygen. Researchers are working on injecting the bacteria with certain cell toxins that would be released once inside the cancerous tumor. Additionally, the bacteria have very strong flagella that help them propel through the tissue to the targeted area. Medication require the presence of networks of blood vessels in helping them travel. If a tumor is found to be very deep within the tissue and not near any blood vessels, it makes it very difficult for the drugs to reach them. However, the Salmonella bacteria do not require blood vessels for their travels. Their flagellar system are like motors that allow them to travel to areas that are embedded deep within tissues. Once inside the tumor, the bacteria are programmed to start producing drugs that would destroy the cancerous cells. L-aribinose triggers the production of such compounds. Salmonella, as well as other bacteria, normally infect their host by deliveing proteins through a molecular needle called a type three secretion system. This molecular needle is what researchers are using to help fight cancer. Through the use of genetic engineering, researchers have modified the protein that the Salmonella bacteria normally secretes, so that it will not harm the host. Instead, the bacteria were engineered to produce a protein known as NY-ESO-1. This protein, which is not found in healthy cells allows the immune system to detect it as foreign and then work to eradicate all of the cells which contain it, mainly the cancer cells. The use of S. typhimurium can greatly aid in the battle against cancer, in ways in which medication failed to do, thus far.
Current Research
In May of 2005, an 18 year old girl, just comming back to Kuwait from India, was found to have abdominal cramps and a high fever. She went for medical help where she had blood drawn for culture. The results of her blood test showed the growth of S. Paratyphi A, and serotyoe of Salmonella enterica. Further testing proved that the mictobe was sensitive to ciprofloxacin. The young girl was prescribed with this medication and told to take it orally two times a day. After consistently taking the medication for one week, the girl still did not show any signs of recovery. She was sent to Infectious Disease Hospital where she experieced a high fever, abdominal pain, and hepatomegaly for ten days. The immediatly tested her for the presence of the malaria parasite, all which came back negative. They provided her with introvenous ceftiaxone, due to her failed response to ciprofloxacin. The patient responded to the new medication within three days and was discharged after nine days with no relapse. This article is important in that it points out how common it is becoming for strains of typhoid salmonella to be resistant to certain medications and thereofore cause the treatment to fail. In 1992 in the United Kingdom, the first case of typhoid fever that was resistant to ciprofloxacin was found. Previously, the strain was found to be resistant to both ciprofloxacin and ceftiaxone, while this case in Kuwait was only resistant to ciprofloxacin. Flouroquinolones are the best way to treat infections by different microbes that cause enteric fevers. The increased resistance to it cuases a major barrier for the medical community. Resistance is hypothesized to be caused by mutations in DNA gyrase. More research needs to be done on the resistance of typhoid salmonella to fluoroquinolones.
References
[Sample reference] Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. "Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney". International Journal of Systematic and Evolutionary Microbiology. 2000. Volume 50. p. 489-500. [1]