Adsorbent chemistries: Difference between revisions
imported>K.H. Wong No edit summary |
imported>K.H. Wong |
||
| (50 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
{{TOC|right}} | |||
Various '''adsorbent chemistries''' in [[chromatography]] are chosen based on the properties of the molecule or [[protein]] of interest and its ability to interact with a stationary phase. [[molecule|Biomolecules]] and proteins in particular can adsorb to a range of solid phases in a selective manner that allows for differential elution and [[purification]]. Adsorbents that have been successfully used in the purification of biomolecules include [[hydrophobic]] [[adsorption|adsorbents]], [[ion]] exchangers and affinity adsorbents. | |||
Biomolecules and proteins in particular can adsorb to a range of solid phases in a selective manner that allows for differential elution and purification. Adsorbents that have been successfully used in the purification of biomolecules include hydrophobic adsorbents, ion exchangers and affinity adsorbents. | |||
==Hydrophobic interaction chromatography== | ==Hydrophobic interaction chromatography== | ||
| Line 10: | Line 8: | ||
===Principles=== | ===Principles=== | ||
Hydrophobic | {{Image|HIC2.jpg|right|450px|Figure 1. Legend: P: Polymer matrix, S: Solute molecule, L: ligand attached to polymer matrix, H: Hydrophobic patch on solute surface, W: Water in bulk solution. Water molecules are more highly ordered at the surface of the hydrophobic ligand and solute than in bulk water. Water molecules shield off the hydrophobic ligand molecules. When salt is added, it interacts strongly with water molecules to leave less water available for L and H to interact with each other.}} | ||
[[Hydrophobic interaction chromatography]] (HIC) is a process that separates molecules based on their [[hydrophobic]]ity, and is typically used as an intermediate step in the [[purification]] of [[proteins]]. The [[hydrophobic]] groups on the surface of molecules will tend to associate with hydrophobic groups on the surface of an insoluble [[matrix]]. The strength of interaction between the [[solute]] molecule and the matrix is related to its [[ion|ionic strength]] and to the ability of the salt ions to alter the orientation of water molecules in the solution. [[Kosmotropic]] salts (which contribute to stability) promote hydrophobic interactions, while [[chaotropic]] salts (which interfere with stability) decrease hydrophobic interactions. Matrices of different binding strengths can be chosen based on the hydrophobic ligands on the surface. Strong hydrophobic matrices will typically have phenyl or octyl groups, and weak hydrophobic matrices will have ether or isopropyl groups. Increasing [[temperature]] tends to strengthen hydrophobic interactions. Compounds that reduce the [[polarity]] of the solvent will decrease hydrophobic interactions. | |||
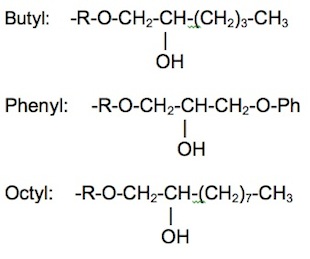
{{Image|HIC.jpg|right|450px|Figure 2. Typical stationary phase ligands used in hydrophobic interaction chromatography.}} | |||
===Procedure=== | |||
Most [[hydrophobic interaction chromatography]] experiments are performed in four main stages: [[equilibrium|equilibration]], sample application and [[wash]], [[elution]], and [[regeneration]]. | |||
( | # Equilibration: The first step is the equilibration of the stationary phase to the desired start conditions, which is done by adding [[salt]] to the mobile phase and passing [[buffer]] through the column. | ||
# Sample application: The next step is sample application and wash. The goal is to bind the target molecule and wash away unbound material. Binding is promoted by moderately high salt concentrations (around 0.5 M). | |||
# Elution: Molecules are released from the hydrophobic surface by a change in the buffer composition, typically by decreasing the salt concentration of the buffer. Other methods such as lowering the temperature, inclusion of organic [[solvents]] or addition of non-ionic detergents can be used to decrease hydrophobic interactions. | |||
# Regeneration: The final step removes all the remaining bound molecules, ensuring that the stationary phase is at full capacity for the next run. | |||
===Applications=== | ===Applications=== | ||
Hydrophobic interaction chromatography can be used to separate proteins and peptides according to their | [[Hydrophobic interaction chromatography]] can be used to separate [[proteins]] and peptides according to their [[hydrophobic]]ity. The procedure is suited as an intermediate step in a multi-step [[purification]] process and is an ideal step after [[ion exchange chromatography]]. For example, [[protein|glycoproteins]] with high carbon content, that often bind irreversibly to saccharide-based [[chromatography|chromatograph]]ic media, can be purified by HIC resins. HIC can be used as an intermediate purification step to separate the correctly folded form of a growth factor, such as IGF-1, from a misfolded, yet stable form. HIC has also been shown to be successful in purifying DNA [[plasmids]]. The Toyopearl Hexyl-650C resin is effective in removing [[endotoxins]] with capacities exceeding 2 million EU/mL of resin, while effectively eliminating [[RNA]] and protein contaminants. Additionally, it is effective in separating the supercoiled and open circular forms of plasmid DNA. | ||
===Reversed Phase Chromatography=== | |||
[[Reversed phase chromatography]] (RPC) separates [[molecule]]s on the basis of [[hydrophobic]]ity. It is closely related to HIC in that both techniques are based on interactions between hydrophobic patches on biomolecule surfaces and the hydrophobic surfaces of a [[chromatography|chromatograph]]ic medium. However, RPC differs in practice in that the surface of an RPC medium is generally highly substituted with [[hydrocarbon]] chains and, thus, more hydrophobic than that of an HIC medium, leading to stronger interactions that must be reversed using non-[[polar]], [[organic]] [[solvents]] for successful [[elution]]. RPC is useful in applications like [[peptide]] mapping or [[purity]] checking. It is often used in the final polishing step of [[oligonucleotide]] or peptide purification. | |||
==Ion exchange chromatography== | ==Ion exchange chromatography== | ||
| Line 33: | Line 34: | ||
===Principles=== | ===Principles=== | ||
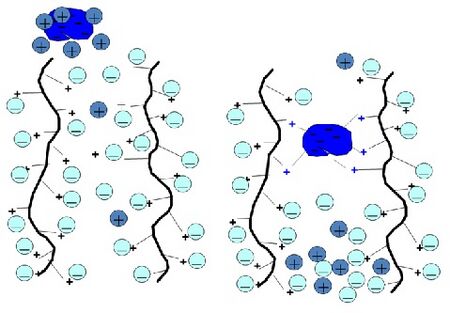
{{Image|Ionexchange.jpg|right|450px|Figure 3. Illustration of an anion exchanger, consisting of a positively-charged resin, that binds to a compound with an overall negative charge. The counter ions, initially in equilibrium with the ion exchanger, are exchanged for the sample compound.}} | |||
[[ | [[Ion exchange chromatography]] is a process that separates [[molecules]] according to their net [[charge]]. A [[column]] that is packed with an insoluble solid phase containing [[ion]]ic groups can reversibly exchange ions with a [[solution]]. Biomolecules attach to ion exchangers through [[electrostatic interactions]] between the molecule's surface charge and the charged groups on the ion exchanger. Some molecules may have both [[anionic]] and [[cationic]] groups. An anion exchanger, which consists of a positively-charged resin, will bind a compound with an overall negative charge. Likewise, a cation exchanger, which consists of a negatively-charged resin, will bind a compound with an overall positive charge. The strength of [[adsorption]] is proportional to the net charge of the molecule. A suitable resin is chosen based on the [[charge]] and [[molecular weight]] of the target molecule or protein. | ||
===Procedure=== | ===Procedure=== | ||
Ion exchange can be carried out in either a column or by a batch procedure. Both methods are carried out in the same definite stages: equilibration of the ion exchanger, addition of sample substances, selective desorption, and regeneration. The ion exchanger is initially in equilibrium with its counter ions. When sample substances enter the ion exchanger bed, the counter ions are exchanged for the sample substances. A gradient of linearly increasing salt concentration is applied after absorption. During desorption, the substance is exchanged for gradient ions and is eluted from the ion exchanger. In the final step, the gradient ions are exchanged for counter ions, thus regenerating the ion exchanger for further use. | [[Ion]] exchange can be carried out in either a [[column]] or by a [[batch]] procedure. Both methods are carried out in the same definite stages: [[equilibrium|equilibration]] of the ion exchanger, addition of sample substances, selective [[desorption]], and [[regeneration]]. The ion exchanger is initially in [[equilibrium]] with its counter ions. When sample substances enter the ion exchanger bed, the counter ions are exchanged for the sample substances. A gradient of linearly increasing [[salt]] concentration is applied after absorption. During desorption, the substance is exchanged for gradient ions and is eluted from the ion exchanger. In the final step, the gradient ions are exchanged for counter ions, thus regenerating the ion exchanger for further use. | ||
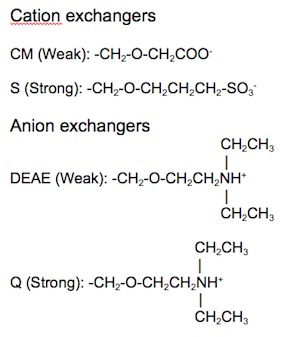
{{Image|IEX2.jpg|right|450px|Figure 4. Chemical structures of typical cation and anion exchangers.}} | |||
There are two methods of desorbing molecules from the ion exchanger: | There are two methods of desorbing molecules from the ion exchanger: | ||
# Varying [[pH]] levels: Changing the pH can reduce the net charges of sample molecules and [[matrix]] [[ligand]]s, thereby influencing selectivity. | |||
# Adding a competing ion: The addition of a competing ion blocks the charges on the ion exchanger and desorbs molecules in order of increasing net charges. | |||
Neutral [[salts]] such as sodium chloride are often used as the desorbing agent because they have little effect on the running pH. | |||
Neutral salts such as sodium chloride are often used as the desorbing agent because they have little effect on the running pH. | |||
Ion exchange matrices can be categorized as being either strong or weak. Strong ion exchange matrices are ionized across a wide range of pH levels, while weak ion exchange matrices are ionized within a narrow range of pH levels. The four most common ion exchangers are shown in Table 1. | Ion exchange matrices can be categorized as being either strong or weak. Strong ion exchange matrices are ionized across a wide range of pH levels, while weak ion exchange matrices are ionized within a narrow range of pH levels. The four most common ion exchangers are shown in Table 1. | ||
{| class="wikitable" | |||
|- | |||
Type of ion exchanger Common | ! Type of ion exchanger | ||
! Common abbreviation | |||
Strong | ! Functional group | ||
Weak | ! pH range for + charge | ||
Strong | ! pH range for - charge | ||
Weak | |- | ||
| Strong anion | |||
| Q | |||
| Quarternary ammonium | |||
| All | |||
| None | |||
|- | |||
| Weak anion | |||
| DEAE | |||
| Diethylaminoethyl | |||
| ≤ 8.5 | |||
| None | |||
|- | |||
| Strong cation | |||
| S | |||
| Sulfonic acid | |||
| None | |||
| > 4 | |||
|- | |||
| Weak cation | |||
| CM | |||
| Carboxymethyl | |||
| None | |||
| All | |||
|} | |||
Table 1. Four common ion exchange chemistries. | |||
==Affinity Chromatography== | ==Affinity Chromatography== | ||
| Line 67: | Line 91: | ||
===Principles=== | ===Principles=== | ||
Affinity chromatography is a method of separating | [[Affinity chromatography]] is a method of separating [[mixture]]s based on highly specific biological interactions, making use of a specific natural or added property in the molecule of interest to isolate it from other molecules in the sample. Examples of such interactions can be found in [[enzymes]] and [[substrates]] or [[receptors]] and [[ligands]]. An affinity ligand specific for the binding site of the target molecule is coupled to a stationary phase, typically a gel [[matrix]] composed of agarose or cross-linked agarose beads. During [[purification]], the specific ligand on the matrix will bind molecules according to its specificity only. The other molecules, which do not possess the defined property, move freely through the column unbound. The bound molecules are subsequently released and [[eluted]] via the addition of a competing substance or by changing the conditions in the column towards [[dissociation]]. | ||
===Adsorbent requirements=== | ===Adsorbent requirements=== | ||
The adsorbents used consist of a matrix to which a ligand with high affinity for the molecules to be separated is reacted with the activated matrix or with a spacer arm attached to the matrix. Agarose beads and cross-linked agarose beads are typical solid packing used as matrices in affinity chromatography. The main requirements for a successful affinity adsorbent are as follows: | The [[adsorbents]] used consist of a [[matrix]] to which a [[ligand]] with high affinity for the molecules to be separated is reacted with the activated matrix or with a spacer arm attached to the matrix. Agarose beads and cross-linked agarose beads are typical solid packing used as matrices in affinity chromatography. The main requirements for a successful affinity adsorbent are as follows: | ||
# The matrix-ligand binding must not affect the ligand’s binding to the target molecule. | |||
# A spacer arm separating the ligand from the matrix should be utilized, especially if the ligand is small. | |||
# Non-specific interactions should be minimized. | |||
# A stable linkage of the ligand to the matrix is needed to ensure proper operation and clean-up for re-use. | |||
===Applications=== | ===Applications=== | ||
A common use of affinity chromatography is for the purification of recombinant proteins. The molecule of interest in solution will have a defined property that can be exploited during the process. For example, there are antibodies in a blood serum sample that are specific for a particular antigenic determinant, which can be isolated through affinity chromatography. The following are steps taken to isolate antibodies in a serum sample using affinity chromatography: | A common use of [[affinity chromatography]] is for the [[purification]] of [[recombinant]] [[proteins]]. The [[molecule]] of interest in solution will have a defined property that can be exploited during the process. For example, there are [[antibodies]] in a [[blood serum]] sample that are specific for a particular [[antigenic determinant]], which can be isolated through affinity chromatography. The following are steps taken to isolate antibodies in a serum sample using affinity chromatography: | ||
# An [[immunoadsorbent]] consisting of a solid matrix to which the antigen is coupled to is prepared. | |||
# Pass the serum over the adsorbent. Antibodies in the mixture specific for the antigen will bind and be retained, while antibodies of other specificities and proteins will pass freely through the column. | |||
# A reagent is used to release the antibodies from the immunoadsorbent. [[Denaturing]] agents like [[urea]] can be used to alter the configuration of the antigen-binding site and break its interaction with the antibody molecule. Buffers with high salt concentration or low pH can disrupt the non-covalent interactions between antibodies and antigen. | |||
# The solution obtained by elution is [[dialysis|dialyzed]] against buffered [[saline]] to remove the elution reagent. | |||
===Immobilized Metal Ion Affinity Chromatography=== | |||
[[Immobilized metal affinity chromatography]] (IMAC) is a commonly used technique in protein purification. IMAC is distinct from other [[affinity chromatography|affinity]] techniques in that it can be used on native [[proteins]] without the need for specialized [[affinity tags]] to facilitate binding. Instead, IMAC relies on the interaction of specific surface [[amino acids]] of the target protein and chelated [[metal]] [[ions]]. Proteins with an affinity for metal ions are retained in the column containing immobilized metal ions, such as cobalt and nickel for the [[purification]] of histidine containing proteins. This allows small differences in protein sequence and structure to be discerned, which is particularly important when a protein of interest is to be separated from a complex mixture. Since many proteins do not naturally have an affinity for metal ions, [[recombinant DNA technology]] can be used to insert a protein tag into the relevant [[gene]]. To [[elute]] the protein of interest, methods such as changing the [[pH]] or adding a competing molecule are used. | |||
==Size exclusion chromatography== | ==Size exclusion chromatography== | ||
| Line 98: | Line 119: | ||
===Principles=== | ===Principles=== | ||
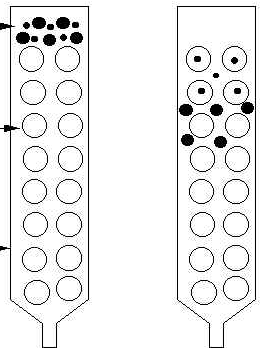
Size exclusion chromatography is a process that separates | [[Size exclusion chromatography]] is a process that separates [[molecule]]s according to their size. This can be done using resins or [[membranes]]. With membranes, the smaller molecules pass through the membrane while larger molecules are held back. Using resins, the larger molecules pass through the column while smaller molecules, which are trapped in the pores of the resin, take longer to flow through. There is no binding between the solid phase and solute molecules. The solid phase is composed of a [[gel]] with calibrated pores; the solids separate by diffusing in and out of these pores. | ||
In [[gel filtration chromatography]], an [[aqueous]] [[solution]] is typically used to transport the sample through the column, while in [[gel permeation chromatography]], an [[organic]] [[solvent]] is used as the mobile phase. Gel-filtration chromatography is used mainly in the fractionation of proteins and water-soluble polymers, while gel-permeation chromatography can be used to determine the molecular weights of organic-soluble polymers. | |||
{{Image|SEC2.png|right|450px|Figure 5. Illustration of gel filtration chromatography. Gel filtration chromatography separates proteins according to their size. Proteins that are small enough enter holes in the beads and travel more slowly than proteins that cannot enter the beads. | |||
}} | |||
===Procedure=== | |||
The following steps are performed during the operation of [[size exclusion chromatography]]: | |||
# [[Gel]] is poured into the [[column]], and a [[buffer]] is run through it to [[equilibrate]] the gel and buffer. | |||
# The sample with possible marker [[solute]] molecules is applied in the same or similar buffer. | |||
# The column is [[eluted]] with the same or similar buffer. | |||
# After fractions are collected, the nonspecifically bound solute molecules are flushed out. | |||
# The column is re-equilibrated for the next run. | |||
===Applications=== | ===Applications=== | ||
[[Size exclusion chromatography]] is a popular method of characterizing biological [[polymers]], such as [[proteins]] and [[nucleic acids]], because it is able to provide good [[molecular weight]] results for polymers; it can be used to determine the [[subunits]] that compose a multimeric protein and to isolate multimers from each other. SEC can also be used to remove small contaminants from protein mixtures and is commonly used as a polishing step in protein [[purification]]. For low pressure operation, a gel medium such as polyacrylamide, dextran or agarose is typically used, while silica or cross-linked polystyrene medium is used for high pressure operation. SEC is not a high [[resolution]] technique and cannot be used to separate components unless they differ by at least two-fold in molecular weight. | |||
Size exclusion chromatography is a popular method of characterizing biological polymers, such as proteins and nucleic acids, because it is able to provide good molecular weight results for polymers. For low pressure operation, a gel medium such as polyacrylamide, dextran or agarose is typically used, while | |||
==References== | ==References== | ||
# Affinity Chromatography: Principles and Methods. Pharmacia Fine Chemicals. | |||
# Hydrophobic Interaction Chromatography. TOSOH Bioscience. http://wolfson.huji.ac.il/purification/PDF/HCIC/TOSOH_HIC.pdf | |||
# Hydrophobic Interaction Chromatography Media Cellufine Butyl, Phenyl & Octyl. Chisso Corporation. http://www.chisso.co.jp/fine/en/cellufine/hydrophobic.html | |||
# Hydrophobic Interaction Chromatography Principles and Methods. Amersham Pharmacia Biotech. | |||
# J. Kennedy, J. Cabral. Recovery Processes for Biological Materials, pp. 426-430. | |||
# Protein Purification Handbook. Amersham Bioscience. | |||
# Sephadex ion exchangers: A guide to ion exchange chromatography. Pharmacia Fine Chemicals. | |||
# W. Lee, K. Lee. Applications of affinity chromatography in proteomics. Analytical Biochemistry, Volume 324, Issue 1, Pages 1-10 (2004). | |||
==Related Articles== | |||
# A. Hardin, C. Harinarayan, G. Malmquist. Ion exchange chromatography of monoclonal antibodies: Effect of resin ligand density on dynamic binding capacity. Journal of Chromatography A, Volume 1216, Issue 20, Pages 4366-4371, (2009). | |||
# C. Grabielle-Madelmont, S. Lesieur, M. Ollivon. Characterization of loaded liposomes by size exclusion chromatography. Journal of Biochemical and Biophysical Methods, Volume 56, Issues 1-3, Pages 189-217 (2003). | |||
# G. Jameson, D. Elmore. Affinity chromatography of bovine trypsin. A rapid separation of bovine α- and β-trypsin. Biochem J. 1974 August; 141(2): 555-565. | |||
Latest revision as of 21:09, 18 December 2010
Various adsorbent chemistries in chromatography are chosen based on the properties of the molecule or protein of interest and its ability to interact with a stationary phase. Biomolecules and proteins in particular can adsorb to a range of solid phases in a selective manner that allows for differential elution and purification. Adsorbents that have been successfully used in the purification of biomolecules include hydrophobic adsorbents, ion exchangers and affinity adsorbents.
Hydrophobic interaction chromatography
Principles
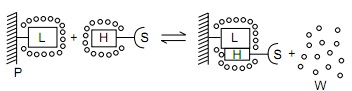
Hydrophobic interaction chromatography (HIC) is a process that separates molecules based on their hydrophobicity, and is typically used as an intermediate step in the purification of proteins. The hydrophobic groups on the surface of molecules will tend to associate with hydrophobic groups on the surface of an insoluble matrix. The strength of interaction between the solute molecule and the matrix is related to its ionic strength and to the ability of the salt ions to alter the orientation of water molecules in the solution. Kosmotropic salts (which contribute to stability) promote hydrophobic interactions, while chaotropic salts (which interfere with stability) decrease hydrophobic interactions. Matrices of different binding strengths can be chosen based on the hydrophobic ligands on the surface. Strong hydrophobic matrices will typically have phenyl or octyl groups, and weak hydrophobic matrices will have ether or isopropyl groups. Increasing temperature tends to strengthen hydrophobic interactions. Compounds that reduce the polarity of the solvent will decrease hydrophobic interactions.
Procedure
Most hydrophobic interaction chromatography experiments are performed in four main stages: equilibration, sample application and wash, elution, and regeneration.
- Equilibration: The first step is the equilibration of the stationary phase to the desired start conditions, which is done by adding salt to the mobile phase and passing buffer through the column.
- Sample application: The next step is sample application and wash. The goal is to bind the target molecule and wash away unbound material. Binding is promoted by moderately high salt concentrations (around 0.5 M).
- Elution: Molecules are released from the hydrophobic surface by a change in the buffer composition, typically by decreasing the salt concentration of the buffer. Other methods such as lowering the temperature, inclusion of organic solvents or addition of non-ionic detergents can be used to decrease hydrophobic interactions.
- Regeneration: The final step removes all the remaining bound molecules, ensuring that the stationary phase is at full capacity for the next run.
Applications
Hydrophobic interaction chromatography can be used to separate proteins and peptides according to their hydrophobicity. The procedure is suited as an intermediate step in a multi-step purification process and is an ideal step after ion exchange chromatography. For example, glycoproteins with high carbon content, that often bind irreversibly to saccharide-based chromatographic media, can be purified by HIC resins. HIC can be used as an intermediate purification step to separate the correctly folded form of a growth factor, such as IGF-1, from a misfolded, yet stable form. HIC has also been shown to be successful in purifying DNA plasmids. The Toyopearl Hexyl-650C resin is effective in removing endotoxins with capacities exceeding 2 million EU/mL of resin, while effectively eliminating RNA and protein contaminants. Additionally, it is effective in separating the supercoiled and open circular forms of plasmid DNA.
Reversed Phase Chromatography
Reversed phase chromatography (RPC) separates molecules on the basis of hydrophobicity. It is closely related to HIC in that both techniques are based on interactions between hydrophobic patches on biomolecule surfaces and the hydrophobic surfaces of a chromatographic medium. However, RPC differs in practice in that the surface of an RPC medium is generally highly substituted with hydrocarbon chains and, thus, more hydrophobic than that of an HIC medium, leading to stronger interactions that must be reversed using non-polar, organic solvents for successful elution. RPC is useful in applications like peptide mapping or purity checking. It is often used in the final polishing step of oligonucleotide or peptide purification.
Ion exchange chromatography
Principles
Ion exchange chromatography is a process that separates molecules according to their net charge. A column that is packed with an insoluble solid phase containing ionic groups can reversibly exchange ions with a solution. Biomolecules attach to ion exchangers through electrostatic interactions between the molecule's surface charge and the charged groups on the ion exchanger. Some molecules may have both anionic and cationic groups. An anion exchanger, which consists of a positively-charged resin, will bind a compound with an overall negative charge. Likewise, a cation exchanger, which consists of a negatively-charged resin, will bind a compound with an overall positive charge. The strength of adsorption is proportional to the net charge of the molecule. A suitable resin is chosen based on the charge and molecular weight of the target molecule or protein.
Procedure
Ion exchange can be carried out in either a column or by a batch procedure. Both methods are carried out in the same definite stages: equilibration of the ion exchanger, addition of sample substances, selective desorption, and regeneration. The ion exchanger is initially in equilibrium with its counter ions. When sample substances enter the ion exchanger bed, the counter ions are exchanged for the sample substances. A gradient of linearly increasing salt concentration is applied after absorption. During desorption, the substance is exchanged for gradient ions and is eluted from the ion exchanger. In the final step, the gradient ions are exchanged for counter ions, thus regenerating the ion exchanger for further use.
There are two methods of desorbing molecules from the ion exchanger:
- Varying pH levels: Changing the pH can reduce the net charges of sample molecules and matrix ligands, thereby influencing selectivity.
- Adding a competing ion: The addition of a competing ion blocks the charges on the ion exchanger and desorbs molecules in order of increasing net charges.
Neutral salts such as sodium chloride are often used as the desorbing agent because they have little effect on the running pH.
Ion exchange matrices can be categorized as being either strong or weak. Strong ion exchange matrices are ionized across a wide range of pH levels, while weak ion exchange matrices are ionized within a narrow range of pH levels. The four most common ion exchangers are shown in Table 1.
| Type of ion exchanger | Common abbreviation | Functional group | pH range for + charge | pH range for - charge |
|---|---|---|---|---|
| Strong anion | Q | Quarternary ammonium | All | None |
| Weak anion | DEAE | Diethylaminoethyl | ≤ 8.5 | None |
| Strong cation | S | Sulfonic acid | None | > 4 |
| Weak cation | CM | Carboxymethyl | None | All |
Table 1. Four common ion exchange chemistries.
Affinity Chromatography
Principles
Affinity chromatography is a method of separating mixtures based on highly specific biological interactions, making use of a specific natural or added property in the molecule of interest to isolate it from other molecules in the sample. Examples of such interactions can be found in enzymes and substrates or receptors and ligands. An affinity ligand specific for the binding site of the target molecule is coupled to a stationary phase, typically a gel matrix composed of agarose or cross-linked agarose beads. During purification, the specific ligand on the matrix will bind molecules according to its specificity only. The other molecules, which do not possess the defined property, move freely through the column unbound. The bound molecules are subsequently released and eluted via the addition of a competing substance or by changing the conditions in the column towards dissociation.
Adsorbent requirements
The adsorbents used consist of a matrix to which a ligand with high affinity for the molecules to be separated is reacted with the activated matrix or with a spacer arm attached to the matrix. Agarose beads and cross-linked agarose beads are typical solid packing used as matrices in affinity chromatography. The main requirements for a successful affinity adsorbent are as follows:
- The matrix-ligand binding must not affect the ligand’s binding to the target molecule.
- A spacer arm separating the ligand from the matrix should be utilized, especially if the ligand is small.
- Non-specific interactions should be minimized.
- A stable linkage of the ligand to the matrix is needed to ensure proper operation and clean-up for re-use.
Applications
A common use of affinity chromatography is for the purification of recombinant proteins. The molecule of interest in solution will have a defined property that can be exploited during the process. For example, there are antibodies in a blood serum sample that are specific for a particular antigenic determinant, which can be isolated through affinity chromatography. The following are steps taken to isolate antibodies in a serum sample using affinity chromatography:
- An immunoadsorbent consisting of a solid matrix to which the antigen is coupled to is prepared.
- Pass the serum over the adsorbent. Antibodies in the mixture specific for the antigen will bind and be retained, while antibodies of other specificities and proteins will pass freely through the column.
- A reagent is used to release the antibodies from the immunoadsorbent. Denaturing agents like urea can be used to alter the configuration of the antigen-binding site and break its interaction with the antibody molecule. Buffers with high salt concentration or low pH can disrupt the non-covalent interactions between antibodies and antigen.
- The solution obtained by elution is dialyzed against buffered saline to remove the elution reagent.
Immobilized Metal Ion Affinity Chromatography
Immobilized metal affinity chromatography (IMAC) is a commonly used technique in protein purification. IMAC is distinct from other affinity techniques in that it can be used on native proteins without the need for specialized affinity tags to facilitate binding. Instead, IMAC relies on the interaction of specific surface amino acids of the target protein and chelated metal ions. Proteins with an affinity for metal ions are retained in the column containing immobilized metal ions, such as cobalt and nickel for the purification of histidine containing proteins. This allows small differences in protein sequence and structure to be discerned, which is particularly important when a protein of interest is to be separated from a complex mixture. Since many proteins do not naturally have an affinity for metal ions, recombinant DNA technology can be used to insert a protein tag into the relevant gene. To elute the protein of interest, methods such as changing the pH or adding a competing molecule are used.
Size exclusion chromatography
Principles
Size exclusion chromatography is a process that separates molecules according to their size. This can be done using resins or membranes. With membranes, the smaller molecules pass through the membrane while larger molecules are held back. Using resins, the larger molecules pass through the column while smaller molecules, which are trapped in the pores of the resin, take longer to flow through. There is no binding between the solid phase and solute molecules. The solid phase is composed of a gel with calibrated pores; the solids separate by diffusing in and out of these pores.
In gel filtration chromatography, an aqueous solution is typically used to transport the sample through the column, while in gel permeation chromatography, an organic solvent is used as the mobile phase. Gel-filtration chromatography is used mainly in the fractionation of proteins and water-soluble polymers, while gel-permeation chromatography can be used to determine the molecular weights of organic-soluble polymers.
Procedure
The following steps are performed during the operation of size exclusion chromatography:
- Gel is poured into the column, and a buffer is run through it to equilibrate the gel and buffer.
- The sample with possible marker solute molecules is applied in the same or similar buffer.
- The column is eluted with the same or similar buffer.
- After fractions are collected, the nonspecifically bound solute molecules are flushed out.
- The column is re-equilibrated for the next run.
Applications
Size exclusion chromatography is a popular method of characterizing biological polymers, such as proteins and nucleic acids, because it is able to provide good molecular weight results for polymers; it can be used to determine the subunits that compose a multimeric protein and to isolate multimers from each other. SEC can also be used to remove small contaminants from protein mixtures and is commonly used as a polishing step in protein purification. For low pressure operation, a gel medium such as polyacrylamide, dextran or agarose is typically used, while silica or cross-linked polystyrene medium is used for high pressure operation. SEC is not a high resolution technique and cannot be used to separate components unless they differ by at least two-fold in molecular weight.
References
- Affinity Chromatography: Principles and Methods. Pharmacia Fine Chemicals.
- Hydrophobic Interaction Chromatography. TOSOH Bioscience. http://wolfson.huji.ac.il/purification/PDF/HCIC/TOSOH_HIC.pdf
- Hydrophobic Interaction Chromatography Media Cellufine Butyl, Phenyl & Octyl. Chisso Corporation. http://www.chisso.co.jp/fine/en/cellufine/hydrophobic.html
- Hydrophobic Interaction Chromatography Principles and Methods. Amersham Pharmacia Biotech.
- J. Kennedy, J. Cabral. Recovery Processes for Biological Materials, pp. 426-430.
- Protein Purification Handbook. Amersham Bioscience.
- Sephadex ion exchangers: A guide to ion exchange chromatography. Pharmacia Fine Chemicals.
- W. Lee, K. Lee. Applications of affinity chromatography in proteomics. Analytical Biochemistry, Volume 324, Issue 1, Pages 1-10 (2004).
Related Articles
- A. Hardin, C. Harinarayan, G. Malmquist. Ion exchange chromatography of monoclonal antibodies: Effect of resin ligand density on dynamic binding capacity. Journal of Chromatography A, Volume 1216, Issue 20, Pages 4366-4371, (2009).
- C. Grabielle-Madelmont, S. Lesieur, M. Ollivon. Characterization of loaded liposomes by size exclusion chromatography. Journal of Biochemical and Biophysical Methods, Volume 56, Issues 1-3, Pages 189-217 (2003).
- G. Jameson, D. Elmore. Affinity chromatography of bovine trypsin. A rapid separation of bovine α- and β-trypsin. Biochem J. 1974 August; 141(2): 555-565.