Halobacterium NRC-1: Difference between revisions
imported>Margaret reinlieb |
mNo edit summary |
||
| (177 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
{{Taxobox | {{Taxobox | ||
| color = pink | | color = pink | ||
| Line 15: | Line 14: | ||
| binomial_authority = | | binomial_authority = | ||
}} | }} | ||
'''''Halobacterium sp. NRC-1''''' is a [[pathogen|non-pathogenic]], [[halophile|halophilic]] [[archaea]] that thrives all over the world in high salt environments, including [[Refineries#Salt production and refining|salt production facilities]], brine inclusions in salt crystals, natural lakes and ponds, and salt marshes. Prior to 1990 ''H. NRC-1'' was classified as an [[archeabacterium]] under the [[prokaryote]] kingdom in the [[two-empire]] system which consisted of [[eukaryote]]s and [[prokaryote]]s. Since 1990 the [[prokaryote]]s were split into [[bacteria]] and [[archaea]] due to their different evolutionary paths and biochemical differences.<ref>{{Cite journal | |||
''Halobacterium sp. NRC-1'' is a halophilic [[archaea]] | |||
| issn = 0146-0749 | | issn = 0146-0749 | ||
| volume = 58 | | volume = 58 | ||
| Line 30: | Line 27: | ||
| url = http://www.ncbi.nlm.nih.gov/pubmed/8177167 | | url = http://www.ncbi.nlm.nih.gov/pubmed/8177167 | ||
}}</ref> | }}</ref> | ||
Like all [[archaea]] ''H. NRC-1'' has no [[nucleus]] or [[organelles]] within the cell, and like other [[archaea]], have evolved many metabolic pathways to allow it to survive in extreme environments.<ref | Like all [[archaea]] ''H. NRC-1'' has no [[nucleus]] or [[organelles]] within the cell, and like other [[archaea]], have evolved many metabolic pathways to allow it to survive in extreme environments.<ref>{{Cite web|last=Waggoner|first=Ben|coauthors=Brian R. Speer|date=20 April 2001|title=Introduction to the Archaea: Life's extremists… |url=http://www.ucmp.berkeley.edu/archaea/archaea.html|publisher=University of California Museum of Paleontology|accessdate=8 November 2013}}</ref> | ||
| title = Archaea | |||
''Halobacterium sp. NRC-1'' is motile using both [[flagella]] and gas vesicles, and respond to their environment by moving toward or away from chemicals in a process called [[chemotaxis]]. Additionally, they can move toward or away from light in a process called [[phototaxis]] by utilizing their sensory [[Bacteriorhodopsin|rhodopsins]]. Phototaxis is an advantageous ability for halobacteria because it enables them to swim away from the high levels of ultra violet and ionizing radiation that they are exposed to on a daily basis. | |||
| url = http:// | ''Halobacterium sp. NRC-1'' reproduce by binary fission and grow best in a 42 degree Celsius, aerobic, high salt environment. | ||
}}</ref> | |||
''Halobacterium sp. NRC-1'' is motile using both [[flagella]] and gas vesicles, and respond to their environment by moving | |||
''Halobacterium sp. NRC-1'' is very easy to culture in the lab and its genome has been completely mapped. Whole-genome [[DNA microarray]]s are available to investigate gene expression. This makes it an excellent model microorganism for research | ''Halobacterium sp. NRC-1'' is very easy to culture in the lab and its genome has been completely mapped and sequenced. Whole-genome [[DNA microarray]]s are available to investigate gene expression. This makes it an excellent model microorganism for research in basic cellular processes, gene expression and as well as for teaching. | ||
==Genome structure== | ==Genome structure== | ||
The genome of ''Halobacterium sp. NRC-1'' was published in 2000. Since that time | The genome of ''Halobacterium sp. NRC-1'' was published in 2000. Since that time scientists have done extensive research to gain insights into both its extremophilic abilities as well as its biological processes. In order to survive in its environment, ''H. NRC-1'' has developed amazing capabilities to repair its own genome, one being a precise base excision repair system. Homologous [[recombination]] plays an important role in [[DNA]] repair as well. Its ability to repair its chromosomes after extensive damage is only exceeded by the extremely radiation resistant [[Deinococcus radiodurans]]bacterium.<ref name=UVRad>{{Cite journal | ||
In order to survive in its | |||
| doi = 10.1186/1746-1448-1-3 | | doi = 10.1186/1746-1448-1-3 | ||
| issn = 1746-1448 | | issn = 1746-1448 | ||
| Line 62: | Line 49: | ||
| date = 2005 | | date = 2005 | ||
| url = http://www.salinesystems.org/content/1/1/3 | | url = http://www.salinesystems.org/content/1/1/3 | ||
}}</ref><ref>{{Cite journal | }}</ref> In fact, ''H. NRC-1'' was found to display a significant number of unique homologs with this bacterium, which suggests that they might have been acquired through [[lateral gene transfer]].<ref name=GenomeSeq>{{Cite journal | ||
| doi = VL - 97 | |||
| volume = 97 | |||
| issue = 22 | |||
| pages = 12176-12181 | |||
| last = Ng | |||
| first = Wailap Victor, et al. | |||
| title = Genome sequence of Halobacterium species NRC-1 | |||
| journal = Proceedings of the National Academy of Sciences of the United States of America | |||
| accessdate = 2009-04-18 | |||
| date = 2000-10-24 | |||
| url = http://www.pnas.org/content/97/22/12176.abstract | |||
}}</ref> | |||
This microorganism also displays other types of defense mechanisms. In combination with its saline environment which provides some protection from UV radiation, the proteins that are produced by ''H. NRC-1'' were found to be highly acidic. This low pH helps the proteins resist the denaturing effects of the high concentration of salts that surrounds it.<ref name=PhysResponse>{{Cite journal | |||
| doi = 10.1007/s00792-005-0437-4 | | doi = 10.1007/s00792-005-0437-4 | ||
| volume = 9 | | volume = 9 | ||
| Line 76: | Line 76: | ||
| url = http://dx.doi.org/10.1007/s00792-005-0437-4 | | url = http://dx.doi.org/10.1007/s00792-005-0437-4 | ||
}}</ref> | }}</ref> | ||
<ref name=PostGenome>{{Cite journal | |||
| doi = 10.1186/1746-1448-2-3 | |||
| doi = 10.1186/1746-1448-3 | |||
| issn = 1746-1448 | | issn = 1746-1448 | ||
| volume = | | volume = 2 | ||
| issue = 1 | | issue = 1 | ||
| pages = | | pages = 3 | ||
| last = | | last = DasSarma | ||
| first = | | first = Shiladitya | ||
| coauthors = Priya DasSarma | | coauthors = Brian Berquist, James Coker, Priya DasSarma, Jochen Muller | ||
| title = | | title = Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1 | ||
| journal = Saline Systems | | journal = Saline Systems | ||
| accessdate = 2009-04-18 | | accessdate = 2009-04-18 | ||
| date = | | date = 2006 | ||
| url = http://www.salinesystems.org/content/ | | url = http://www.salinesystems.org/content/2/1/3 | ||
}} </ref> | }}</ref> | ||
[[ | |||
| | ''Halobacterium sp. NRC-1'' contains the smallest [[genome]] to date among the halophiles.<ref name=PostGenome/> | ||
| volume = | It is 2,571,010 bp in size, and is composed of a large GC-rich chromosome (2,014,239 bp, 68 % G+C), and two smaller extrachromosomal [[replicon]]s, pNRC100 (191,346 bp) and pNRC200 (365,425 bp), with 58–59 % G+C composition. The large chromosome contains 2,111 genes, pNRC200 contains 374, and pNRC100 contains 197. Of the 2,682 genes in the genome, only 1,658 coded for proteins that had significant matches to the [[genome database]]. A significantly smaller fraction of the genes on pNRC200 and pNRC100 matched to genes of known function in the databases. The genes on the large chromosome had a 45% match rate, while pNRC200 had a 32% match rate and pNRC100 had only 26% of its genome match up to genes of known function. 52 [[RNA]] genes have also been identified. Interestingly, about 40 genes that are located on the small [[replicons]], pNRC100 and pNRC200, code for functions likely to be essential or important for cell livelihood (e.g. [[thioredoxin]] and [[thioredoxin reductase]], a [[cytochrome oxidase]], a [[DNA]] [[polymerase]], multiple [[TATA]]-binding proteins (TBP) and transcription factor B (TFB) transcription factors, and the only [[arginyl-tRNA synthetase]] in the genome. In fact, these two [[replicon]]s share 145,428 base pairs of identical DNA.<ref name=GenomeSeq/> | ||
| issue = | |||
| pages = | After the publication of ''Halobacterium sp. NRC-1''’s complete genome, the mapping of another closely related microorganism, ''[[Halobacterium H. Salinarum]]'', was published as well. Interestingly, genomic comparisons between these two [[halophiles]] showed that the large chromosomes were virtually identical to one another while the smaller [[replicons]] carried different genes.<ref name=ModelOrganism>{{Cite journal | ||
| last = | | doi = 10.1099/mic.0.28504-0 | ||
| first = | | volume = 152 | ||
| title = | | issue = 3 | ||
| journal = | | pages = 585-590 | ||
| accessdate = 2009-04- | | last = Soppa | ||
| date = | | first = Jorg | ||
| url = http:// | | title = From genomes to function: haloarchaea as model organisms | ||
| journal = Microbiology | |||
| accessdate = 2009-04-18 | |||
| date = 2006-03-01 | |||
| url = http://mic.sgmjournals.org/cgi/content/abstract/152/3/585 | |||
}}</ref> | }}</ref> | ||
It has been extensively | This information has cast new light on how the species differ from each other and the role of [[replicons]] in these organisms. While much of the published literature on ''Halobacterium sp. NRC-1'' refers to the smaller genetic elements as replicons or megaplasmids, their characteristics don't really fit the definition of these terms. [[Replicon]]s are considered to be exact copies of specific sequences of an original DNA or RNA genome, or even a whole copy of the original genome. Plasmids are referred to as being small extra chromosomal [[DNA]] elements that carry relatively few genes that code for genetic information that is not essential to an organism's biological processes. Considering that the genetic information carried on these [[replicon]]s not only code for information that is essential to the organism's survival, but also contain nucleotide sequences that are not identical to that of its larger chromosome, scientists are beginning to refer to them as "minichromosomes" rather than megaplasmids or replicons.<ref name=PostGenome/> | ||
==Cell structure and metabolism== | |||
''Halobacterium sp.NRC-1'' is a rod shaped [[aerobic]] [[chemoorganotroph]] which uses organic molecules as its source of energy, carbon and electrons. This halophile possesses [[facultative anaerobic]] and phototrophic capabilities as well. Research has shown that it is unable to metabolize sugars, but instead rely on amino acids that are eventually [[catabolize]]d by the citric acid cycle during [[aerobic respiration]]. It survives on the remains of less halophilic organisms that have [[lysed]] due to the overwhelming amounts of salt in the environment. Outside of their natural environment, cells are cultured best in a [[complex medium]]. A [[minimal medium]] described for ''Halobacterium'' includes all but 5 of the 20 [[amino acid]]s for growth.<ref name=GenomeSeq/> | |||
This microorganism has been studied extensively and has been shown to contain some of the usual features found in halophilic archaea, for example, an [[S-layer glycoprotein]], [[ether-linked lipids]], and [[purple membrane]].<ref name=UnderstandAdapt>{{Cite journal | |||
| doi = 10.1101/gr.190201 | | doi = 10.1101/gr.190201 | ||
| issn = 1088-9051 | | issn = 1088-9051 | ||
| Line 136: | Line 128: | ||
| url = http://www.ncbi.nlm.nih.gov/pubmed/11591641 | | url = http://www.ncbi.nlm.nih.gov/pubmed/11591641 | ||
}}</ref> | }}</ref> | ||
This purple membrane consists of the light-driven ion transporters [[bacteriorhodopsin]] and [[halorhodopsin]], and the [[phototaxis]] receptors, sensory [[rhodopsin]]s I and II.<ref name=GenomeSeq/> In order to survive in low oxygen environments, ''Halobacterium sp. NRC-1'' increases its production of [[Bacteriorhodopsin]], which is a unique protein that can use light as an energy source, much like [[chlorophyll]] can in [[cyanobacteria]] and phototrophic [[eukaryotes]]. When the [[retinal]] in [[Bacteriorhodopsin]] absorbs light, it results in a series of conformational changes that translocates protons through the cell's membrane into the [[periplasmic]] space. This light driven proton pumping generates an electrochemical proton gradient which is then used to power the synthesis of [[ATP]]. This phototrophic capability is particularly useful to ''Halobacterium sp. NRC-1'' as oxygen is not very soluble in concentrated salt solutions. In addition to its ability to use light as an energy source, it is also capable of anaerobic respiration using [[dimethyl sulfoxide]] ([[DMSO]]) and [[trimethylamine-N-oxide]] ([[TMAO]]) as terminal electron acceptors. [[Arginine]] [[fermentation]] can also be used for anaerobic energy production as well.<ref name=GeneAnaerobic>{{Cite journal | |||
| doi = 10.1128/JB.187.5.1659-1667.2005 | | doi = 10.1128/JB.187.5.1659-1667.2005 | ||
| volume = 187 | | volume = 187 | ||
| Line 152: | Line 143: | ||
}}</ref> | }}</ref> | ||
''Halobacterium sp. NRC-1'' is also classified as an obligate [[halophilic]] microorganism which has adapted to be able to grow in conditions of extremely high salinity, up to 10 times that of seawater.<ref name=GenomeSeq/> | |||
In order to survive under these conditions it maintains a very high concentration of salts internally in the form of KCl to enable it to remain isotonic to its preferred environment.<ref>{{Cite journal | |||
''Halobacterium NRC-1'' is an | | doi = 10.1186/1746-1448-3-6 | ||
| issn = 1746-1448 | |||
| volume = 3 | |||
| issue = 1 | |||
| pages = 6 | |||
| last = Coker | |||
| first = James | |||
| coauthors = Priya DasSarma, Jeffrey Kumar, Jochen Muller, Shiladitya DasSarma | |||
| title = Transcriptional profiling of the model Archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature | |||
| journal = Saline Systems | |||
| accessdate = 2009-04-18 | |||
| date = 2007 | |||
| url = http://www.salinesystems.org/content/3/1/6 | |||
}} </ref> | |||
[[Halorhodopsin]] plays a very important role in helping to maintain the osmotic balance within the cell. This membrane protein acts as a light driven pump by transporting chloride and potassium ions into the cell. [[Halorhodopsin]] saves the organism a large amount of metabolic energy by using the energy of the yellow light that it captures to fuel the movement of these ions.<ref name=Halorhodopsin>{{Cite journal | |||
| issn = 0959-440X | |||
| volume = 8 | |||
| issue = 4 | |||
| pages = 489-500 | |||
| last = Oesterhelt | |||
| first = D | |||
| title = The structure and mechanism of the family of retinal proteins from halophilic archaea | |||
| journal = Current Opinion in Structural Biology | |||
| accessdate = 2009-04-26 | |||
| date = 1998-08 | |||
| url = http://www.ncbi.nlm.nih.gov/pubmed/9729742 | |||
}}</ref> | |||
==Ecology== | ==Ecology== | ||
[[Image: ISS007-E-13002.jpg|right|thumb|350px|-Image ISS007-E-13002 from the ISS of the Great Salt Lake showing purple tint due to ''halobacterium'' presence and increased [[bacteriorhodopsin]] production due to high salt concentration. Image courtesy of the Image Science & Analysis Laboratory, NASA Johnson Space Center]] | [[Image: ISS007-E-13002.jpg|right|thumb|350px|-Image ISS007-E-13002 from the ISS of the Great Salt Lake showing purple tint due to ''halobacterium'' presence and increased [[bacteriorhodopsin]] production due to high salt concentration. Image courtesy of the Image Science & Analysis Laboratory, NASA Johnson Space Center]] | ||
''Halobacterium | ''Halobacterium NRC-1'' is one of many strains of ''halobacterium'' which thrive in extremely high salinity environments such as salt lakes, salt marshes and salt drying ponds. Their optimal temperature for reproduction is 42°C. Often these highly saline bodies of water will be tinted red or purple. It is the red/purple color of the [[bacteriorhodopsin]]s that give the purple color you often see in these highly saline environments. Bacteriorhodopsin consists of a photosensitive protein pigment called [[retinal]]. This protein pigment is responsible for NRC-1's colorful properties. The more saline the environment the more colorful the water will be because ''halobacterium'' increase their production of [[bacteriorhodopsin]] in response to drops in oxygen, which is less soluble in saline solutions. | ||
There are not many other organisms that can survive in these high salt environments, in fact one of its primary sources of food is the [[amino acid]]s of other organisms which have [[lyse]]d due to the high salt concentration in this environment. Brine shrimp are one of a few other organisms that can survive the high salt concentration, and they feed almost exclusively on the [[halobacteria]]. In addition to [[rhodopsin]], ''Halobacterium NRC-1'' produce [[carotenoid]]s, which are red organic pigments that can also serve as antioxidants. The flamingo, whose pink color comes exclusively from the [[carotenoid]]s in its diet (it doesn't have the ability to produce these pigments naturally) feed on the brine shrimp (who are capable of producing these pigments). These same brine shrimp have fed on the [[carotenoid]] producing [[halobacteria]]. So interestingly enough, these small organisms, the brine shrimp and the [[halobacteria]], are the ones responsible for the flamingos beautiful pink coloring. | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
''Halobacterium sp. NRC-1'' was one of the first [[Archaea]] to have its genome fully mapped and published. | ''Halobacterium NRC-1'' can easily be analyzed just by placing it in a [[hypotonic]] solution. Due to the higher salt concentration within the microorganism in comparison to its less salinic environment, water will flow into the cell following its concentration gradient, causing the cell to swell and undergo lysis. This explosion releases this microorganism's proteins, which then can be used by researchers for genetic analysis. ''Halobacterium sp. NRC-1'' was one of the first [[Archaea]] to have its genome fully mapped and published. Since that time several more [[archaea]] have been successfully mapped and a few others partially mapped. This allows scientists to analyze the properties of halobacterium [[in silico]] to determine the activity of genes. Transformation tools have also been developed to create loss-of-function mutants which aid in research. Mutant strains of ''Halobacterium sp. NRC-1'' are experimentally designed to carry inoperative [[knockout genes]] which alter this organism's normal cellular processes. Scientists can then infer the function of the mutated gene by comparing the physical and biochemical characteristics of the original ''Halobacterium sp. NRC-1'' with the characteristics of the new mutant. These gene knockouts are making it possible to analyze this organism's ability to withstand extreme environmental conditions, including high levels of radiation, severe temperature changes, and fluctuations in oxygen levels. <ref name=ModelOrganism/> | ||
The ability of this organism to be easily cultured along with these advanced research tools make ''Halobacterium sp. NRC-1'' an ideal model for the testing of gene functions. | |||
In addition | In addition, ''Halobacterium sp. NRC-1'' is an ideal study tool because it is an [[extremophile]], it exists in environments that are very unfriendly toward life. This makes it very useful for studying many biological questions especially those involved with adaptation and survival in extreme environments. In fact, much research is being done to investigate whether or not ''halobacterium'' could be potential candidates for extraterrestrial life, such as on Mars or Europa. Recent research has shown that ''Halobacterium sp. NRC-1'' can not only survive at temperatures far below its optimal growth temperature, but continue to reproduce as well.<ref name=ExtremeHalo>{{citation | ||
| contribution =Survival and growth of Halobacterium sp. NRC-1 following incubation at -15°C, freezing or freeze-drying, and the protective effect of cations | | contribution =Survival and growth of Halobacterium sp. NRC-1 following incubation at -15°C, freezing or freeze-drying, and the protective effect of cations | ||
| volume = 545 | | volume = 545 | ||
| Line 190: | Line 197: | ||
==Current Research== | ==Current Research== | ||
===Protective effect of cations in extreme environments=== | |||
The [[extremophillic]] properties and biological responses of ''Halobacteruim NRC-1'' are currently being investigated to test the organism's survival potential in the harsh environmental conditions that exist on Mars. The recent discoveries that [[meterorites]] from Mars contain a mineral form of sodium chloride as well as the isolation of halophilic [[archaea]] from ancient rock salt has led scientists to believe that the existence of ''Halobacterium sp. NRC-1'' in a Martian environment is quite possible. ''Halobacterium'' are known to be slightly [[thermophilic]] organisms, showing optimum growth at 40 degrees [[Celsius]]. In this experiment, cells of ''Halobacterium sp. NRC-1'' were freeze dried and suspended in buffers containing specific cations, including Mg++,Ca++,or K+. Results showed that ''NRC-1'' was still capable of slow, yet steady growth at the extremely low temperature of -15 degrees [[Celsius]]. All of the cations appeared to show protective qualities during the freeze-drying, with K+ being the most effective. The conditions of Martian environment include very low temperatures and low water availability. Scientists will continue to research ''NRC-1''’s ability to survive in such inhospitable environments such as the one on Mars. Hopefully, these future findings will provide us some further insight in our search for life beyond our planet.<ref name=ExtremeHalo/> | |||
==== | ===Radiation resistance=== | ||
''Halobacterium sp. NRC-1'' is known to be extremely radiation resistant due to protection from its pigments, the saline environment it thrives in, and its ability to repair its [[DNA]] after extensive damage. In this study the scientists wanted to find out if they could produce a mutant strain that would be even more resistant to ionizing radiation, and determine what genes and mechanisms were responsible for providing the increased resistance. To do this, they exposed cultures of ''H. NRC-1'' to four cycles of irradiation with high doses of 18-20 MeV (mega-electron volt) electrons from a standard medical LINAC (particle accelerator), allowing the cultures to recover between doses. At the end of this process they were able to isolate two mutant strains (named LH5 and LH7a) with an LD<sub>50</sub> (median lethal dose) of greater than 11 kGy (kilo-Grays) (standard measure of absorbed radiation dose) which makes them the most radiation resistant organism known, even more resistant than [[Deinococcus radiodurans]] (LD<sub>50</sub> 7.9 kGy). The wild-type ''H. NRC-1'' was found to have an LD<sub>50</sub> of 5.4 kGy while the mutant strains were 11.9 kGy for LH5 and 12.1 kGy for LH7a. Unlike most other microbes like [[Deinococcus radiodurans|D. radiodurans]] which show the most radiation resistance during their stationary phase, ''H. NRC-1'' is most resistant when in its growth phase. Tests verified that this was true for the mutants as well. They were also able to rule out a slowdown in growth rate, which is one of the methods employed by [[Deinococcus radiodurans|D. radiodurans]], as the reason for the increased resistance. To do this, they compared the doubling rate of the mutant strains with that of the wild-type ''H. NRC-1'' and found it to be the same. They then used whole-genome [[transcriptome]] analysis to compare the mutant strains with the wild type ''H. NRC-1'' to determine what had given the mutant strains their increased radiation resistance. They found significant up-regulation of an [[operon]] containing two single-stranded DNA-binding protein (RPA-replication protein A) genes, VNG2160 (rfa3) and VNG2162, plus a third gene, VNG2163, which is unknown. While the authors tests did not identify the actual role this [[operon]] plays in providing the increased resistance to radiation, they propose that it does facilitate the [[DNA]] repair machinery and/or protects the repair intermediates to maximize radiation resistance.<ref>{{Cite journal | |||
| doi = 10.1667/RR0935.1 | |||
| volume = 168 | |||
| issue = 4 | |||
| pages = 507-514 | |||
| last = Linda C. DeVeaux | |||
| coauthors = Jochen A. Müller, Jonathon Smith, Jill Petrisko, Douglas P. Wells, Shiladitya DasSarma | |||
| title = Extremely Radiation-Resistant Mutants of a Halophilic Archaeon with Increased Single-Stranded DNA-Binding Protein (RPA) Gene Expression | |||
| journal = Radiation Research | |||
| accessdate = 2009-05-10 | |||
| date = 2007 | |||
| url = http://www.bioone.org/doi/abs/10.1667/RR0935.1 | |||
}}</ref> | |||
==== | ===Peptide delivery vehicle for biotechnology=== | ||
''Halobacterium sp. NRC-1'' is also playing a significant role in the advancement of immunological biotechnology. Recent experimental findings show that recombinant gas vesicles from a mutant strain of ''Halobacterium sp. NRC-1'' have the potential to serve as [[antigen]] display/delivery systems. [[DNA]] segments of an [[SIV]] ([[simian]] [[immunodeficiency]] [[virus]]) gene were inserted into a specific site on a GvpC gene (gas vesicle protein C) in the ''Halobacterium''. The gene was successfully taken up by the microorganism and recombination had been accomplished. Subsequently, the gas vesicles began to express the new recombinant/pathogenic proteins at their surface. Mice were then immunized with these antigen presenting [[SIV]] recombinant gas vesicles. [[Antibody]] production was monitored, and an increased level of [[humoral]] response was observed. After a 12 week period, a booster shot was given. 43 weeks after the booster shot was administered, an elevated number of antibodies still remained and was recorded. Research showed that this increased production of antibodies was long lived even in the absence of an [[adjuvant]], and immunologic memory had been proven. These results indicate that the gas vesicles of ''Halobacterium sp. NRC-1'' show great potential in their possible roles as immunizing agents.<ref name=Recombinant>{{Cite journal | |||
| doi = 10.1186/1472-6750-8-9 | |||
| volume = 8 | |||
| pages = 9 | |||
| last = Sremac | |||
| first = Marinko | |||
| coauthors = Elizabeth S Stuart | |||
| title = Recombinant gas vesicles from Halobacterium sp. displaying SIV peptides demonstrate biotechnology potential as a pathogen peptide delivery vehicle | |||
| doi = 10. | | journal = BMC Biotechnology | ||
| volume = | | accessdate = 2009-04-30 | ||
| date = 2008 | |||
| pages = | | url = http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2270826 | ||
| last = | |||
| first = | |||
| coauthors = | |||
| title = | |||
| journal = | |||
| accessdate = 2009-04- | |||
| date = | |||
| url = http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid= | |||
}}</ref> | }}</ref> | ||
==References== | ==References== | ||
{{reflist| | {{reflist|2}}[[Category:Suggestion Bot Tag]] | ||
Latest revision as of 11:00, 25 August 2024
| Halobacterium sp. NRC-1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Halobacterium sp. NRC-1 |
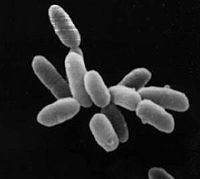
Halobacterium sp. NRC-1 is a non-pathogenic, halophilic archaea that thrives all over the world in high salt environments, including salt production facilities, brine inclusions in salt crystals, natural lakes and ponds, and salt marshes. Prior to 1990 H. NRC-1 was classified as an archeabacterium under the prokaryote kingdom in the two-empire system which consisted of eukaryotes and prokaryotes. Since 1990 the prokaryotes were split into bacteria and archaea due to their different evolutionary paths and biochemical differences.[1] Like all archaea H. NRC-1 has no nucleus or organelles within the cell, and like other archaea, have evolved many metabolic pathways to allow it to survive in extreme environments.[2]
Halobacterium sp. NRC-1 is motile using both flagella and gas vesicles, and respond to their environment by moving toward or away from chemicals in a process called chemotaxis. Additionally, they can move toward or away from light in a process called phototaxis by utilizing their sensory rhodopsins. Phototaxis is an advantageous ability for halobacteria because it enables them to swim away from the high levels of ultra violet and ionizing radiation that they are exposed to on a daily basis. Halobacterium sp. NRC-1 reproduce by binary fission and grow best in a 42 degree Celsius, aerobic, high salt environment.
Halobacterium sp. NRC-1 is very easy to culture in the lab and its genome has been completely mapped and sequenced. Whole-genome DNA microarrays are available to investigate gene expression. This makes it an excellent model microorganism for research in basic cellular processes, gene expression and as well as for teaching.
Genome structure
The genome of Halobacterium sp. NRC-1 was published in 2000. Since that time scientists have done extensive research to gain insights into both its extremophilic abilities as well as its biological processes. In order to survive in its environment, H. NRC-1 has developed amazing capabilities to repair its own genome, one being a precise base excision repair system. Homologous recombination plays an important role in DNA repair as well. Its ability to repair its chromosomes after extensive damage is only exceeded by the extremely radiation resistant Deinococcus radioduransbacterium.[3] In fact, H. NRC-1 was found to display a significant number of unique homologs with this bacterium, which suggests that they might have been acquired through lateral gene transfer.[4] This microorganism also displays other types of defense mechanisms. In combination with its saline environment which provides some protection from UV radiation, the proteins that are produced by H. NRC-1 were found to be highly acidic. This low pH helps the proteins resist the denaturing effects of the high concentration of salts that surrounds it.[5] [6]
Halobacterium sp. NRC-1 contains the smallest genome to date among the halophiles.[6] It is 2,571,010 bp in size, and is composed of a large GC-rich chromosome (2,014,239 bp, 68 % G+C), and two smaller extrachromosomal replicons, pNRC100 (191,346 bp) and pNRC200 (365,425 bp), with 58–59 % G+C composition. The large chromosome contains 2,111 genes, pNRC200 contains 374, and pNRC100 contains 197. Of the 2,682 genes in the genome, only 1,658 coded for proteins that had significant matches to the genome database. A significantly smaller fraction of the genes on pNRC200 and pNRC100 matched to genes of known function in the databases. The genes on the large chromosome had a 45% match rate, while pNRC200 had a 32% match rate and pNRC100 had only 26% of its genome match up to genes of known function. 52 RNA genes have also been identified. Interestingly, about 40 genes that are located on the small replicons, pNRC100 and pNRC200, code for functions likely to be essential or important for cell livelihood (e.g. thioredoxin and thioredoxin reductase, a cytochrome oxidase, a DNA polymerase, multiple TATA-binding proteins (TBP) and transcription factor B (TFB) transcription factors, and the only arginyl-tRNA synthetase in the genome. In fact, these two replicons share 145,428 base pairs of identical DNA.[4]
After the publication of Halobacterium sp. NRC-1’s complete genome, the mapping of another closely related microorganism, Halobacterium H. Salinarum, was published as well. Interestingly, genomic comparisons between these two halophiles showed that the large chromosomes were virtually identical to one another while the smaller replicons carried different genes.[7] This information has cast new light on how the species differ from each other and the role of replicons in these organisms. While much of the published literature on Halobacterium sp. NRC-1 refers to the smaller genetic elements as replicons or megaplasmids, their characteristics don't really fit the definition of these terms. Replicons are considered to be exact copies of specific sequences of an original DNA or RNA genome, or even a whole copy of the original genome. Plasmids are referred to as being small extra chromosomal DNA elements that carry relatively few genes that code for genetic information that is not essential to an organism's biological processes. Considering that the genetic information carried on these replicons not only code for information that is essential to the organism's survival, but also contain nucleotide sequences that are not identical to that of its larger chromosome, scientists are beginning to refer to them as "minichromosomes" rather than megaplasmids or replicons.[6]
Cell structure and metabolism
Halobacterium sp.NRC-1 is a rod shaped aerobic chemoorganotroph which uses organic molecules as its source of energy, carbon and electrons. This halophile possesses facultative anaerobic and phototrophic capabilities as well. Research has shown that it is unable to metabolize sugars, but instead rely on amino acids that are eventually catabolized by the citric acid cycle during aerobic respiration. It survives on the remains of less halophilic organisms that have lysed due to the overwhelming amounts of salt in the environment. Outside of their natural environment, cells are cultured best in a complex medium. A minimal medium described for Halobacterium includes all but 5 of the 20 amino acids for growth.[4]
This microorganism has been studied extensively and has been shown to contain some of the usual features found in halophilic archaea, for example, an S-layer glycoprotein, ether-linked lipids, and purple membrane.[8] This purple membrane consists of the light-driven ion transporters bacteriorhodopsin and halorhodopsin, and the phototaxis receptors, sensory rhodopsins I and II.[4] In order to survive in low oxygen environments, Halobacterium sp. NRC-1 increases its production of Bacteriorhodopsin, which is a unique protein that can use light as an energy source, much like chlorophyll can in cyanobacteria and phototrophic eukaryotes. When the retinal in Bacteriorhodopsin absorbs light, it results in a series of conformational changes that translocates protons through the cell's membrane into the periplasmic space. This light driven proton pumping generates an electrochemical proton gradient which is then used to power the synthesis of ATP. This phototrophic capability is particularly useful to Halobacterium sp. NRC-1 as oxygen is not very soluble in concentrated salt solutions. In addition to its ability to use light as an energy source, it is also capable of anaerobic respiration using dimethyl sulfoxide (DMSO) and trimethylamine-N-oxide (TMAO) as terminal electron acceptors. Arginine fermentation can also be used for anaerobic energy production as well.[9]
Halobacterium sp. NRC-1 is also classified as an obligate halophilic microorganism which has adapted to be able to grow in conditions of extremely high salinity, up to 10 times that of seawater.[4] In order to survive under these conditions it maintains a very high concentration of salts internally in the form of KCl to enable it to remain isotonic to its preferred environment.[10] Halorhodopsin plays a very important role in helping to maintain the osmotic balance within the cell. This membrane protein acts as a light driven pump by transporting chloride and potassium ions into the cell. Halorhodopsin saves the organism a large amount of metabolic energy by using the energy of the yellow light that it captures to fuel the movement of these ions.[11]
Ecology
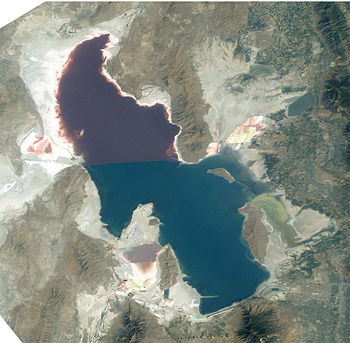
Halobacterium NRC-1 is one of many strains of halobacterium which thrive in extremely high salinity environments such as salt lakes, salt marshes and salt drying ponds. Their optimal temperature for reproduction is 42°C. Often these highly saline bodies of water will be tinted red or purple. It is the red/purple color of the bacteriorhodopsins that give the purple color you often see in these highly saline environments. Bacteriorhodopsin consists of a photosensitive protein pigment called retinal. This protein pigment is responsible for NRC-1's colorful properties. The more saline the environment the more colorful the water will be because halobacterium increase their production of bacteriorhodopsin in response to drops in oxygen, which is less soluble in saline solutions.
There are not many other organisms that can survive in these high salt environments, in fact one of its primary sources of food is the amino acids of other organisms which have lysed due to the high salt concentration in this environment. Brine shrimp are one of a few other organisms that can survive the high salt concentration, and they feed almost exclusively on the halobacteria. In addition to rhodopsin, Halobacterium NRC-1 produce carotenoids, which are red organic pigments that can also serve as antioxidants. The flamingo, whose pink color comes exclusively from the carotenoids in its diet (it doesn't have the ability to produce these pigments naturally) feed on the brine shrimp (who are capable of producing these pigments). These same brine shrimp have fed on the carotenoid producing halobacteria. So interestingly enough, these small organisms, the brine shrimp and the halobacteria, are the ones responsible for the flamingos beautiful pink coloring.
Application to Biotechnology
Halobacterium NRC-1 can easily be analyzed just by placing it in a hypotonic solution. Due to the higher salt concentration within the microorganism in comparison to its less salinic environment, water will flow into the cell following its concentration gradient, causing the cell to swell and undergo lysis. This explosion releases this microorganism's proteins, which then can be used by researchers for genetic analysis. Halobacterium sp. NRC-1 was one of the first Archaea to have its genome fully mapped and published. Since that time several more archaea have been successfully mapped and a few others partially mapped. This allows scientists to analyze the properties of halobacterium in silico to determine the activity of genes. Transformation tools have also been developed to create loss-of-function mutants which aid in research. Mutant strains of Halobacterium sp. NRC-1 are experimentally designed to carry inoperative knockout genes which alter this organism's normal cellular processes. Scientists can then infer the function of the mutated gene by comparing the physical and biochemical characteristics of the original Halobacterium sp. NRC-1 with the characteristics of the new mutant. These gene knockouts are making it possible to analyze this organism's ability to withstand extreme environmental conditions, including high levels of radiation, severe temperature changes, and fluctuations in oxygen levels. [7] The ability of this organism to be easily cultured along with these advanced research tools make Halobacterium sp. NRC-1 an ideal model for the testing of gene functions.
In addition, Halobacterium sp. NRC-1 is an ideal study tool because it is an extremophile, it exists in environments that are very unfriendly toward life. This makes it very useful for studying many biological questions especially those involved with adaptation and survival in extreme environments. In fact, much research is being done to investigate whether or not halobacterium could be potential candidates for extraterrestrial life, such as on Mars or Europa. Recent research has shown that Halobacterium sp. NRC-1 can not only survive at temperatures far below its optimal growth temperature, but continue to reproduce as well.[12]
Current Research
Protective effect of cations in extreme environments
The extremophillic properties and biological responses of Halobacteruim NRC-1 are currently being investigated to test the organism's survival potential in the harsh environmental conditions that exist on Mars. The recent discoveries that meterorites from Mars contain a mineral form of sodium chloride as well as the isolation of halophilic archaea from ancient rock salt has led scientists to believe that the existence of Halobacterium sp. NRC-1 in a Martian environment is quite possible. Halobacterium are known to be slightly thermophilic organisms, showing optimum growth at 40 degrees Celsius. In this experiment, cells of Halobacterium sp. NRC-1 were freeze dried and suspended in buffers containing specific cations, including Mg++,Ca++,or K+. Results showed that NRC-1 was still capable of slow, yet steady growth at the extremely low temperature of -15 degrees Celsius. All of the cations appeared to show protective qualities during the freeze-drying, with K+ being the most effective. The conditions of Martian environment include very low temperatures and low water availability. Scientists will continue to research NRC-1’s ability to survive in such inhospitable environments such as the one on Mars. Hopefully, these future findings will provide us some further insight in our search for life beyond our planet.[12]
Radiation resistance
Halobacterium sp. NRC-1 is known to be extremely radiation resistant due to protection from its pigments, the saline environment it thrives in, and its ability to repair its DNA after extensive damage. In this study the scientists wanted to find out if they could produce a mutant strain that would be even more resistant to ionizing radiation, and determine what genes and mechanisms were responsible for providing the increased resistance. To do this, they exposed cultures of H. NRC-1 to four cycles of irradiation with high doses of 18-20 MeV (mega-electron volt) electrons from a standard medical LINAC (particle accelerator), allowing the cultures to recover between doses. At the end of this process they were able to isolate two mutant strains (named LH5 and LH7a) with an LD50 (median lethal dose) of greater than 11 kGy (kilo-Grays) (standard measure of absorbed radiation dose) which makes them the most radiation resistant organism known, even more resistant than Deinococcus radiodurans (LD50 7.9 kGy). The wild-type H. NRC-1 was found to have an LD50 of 5.4 kGy while the mutant strains were 11.9 kGy for LH5 and 12.1 kGy for LH7a. Unlike most other microbes like D. radiodurans which show the most radiation resistance during their stationary phase, H. NRC-1 is most resistant when in its growth phase. Tests verified that this was true for the mutants as well. They were also able to rule out a slowdown in growth rate, which is one of the methods employed by D. radiodurans, as the reason for the increased resistance. To do this, they compared the doubling rate of the mutant strains with that of the wild-type H. NRC-1 and found it to be the same. They then used whole-genome transcriptome analysis to compare the mutant strains with the wild type H. NRC-1 to determine what had given the mutant strains their increased radiation resistance. They found significant up-regulation of an operon containing two single-stranded DNA-binding protein (RPA-replication protein A) genes, VNG2160 (rfa3) and VNG2162, plus a third gene, VNG2163, which is unknown. While the authors tests did not identify the actual role this operon plays in providing the increased resistance to radiation, they propose that it does facilitate the DNA repair machinery and/or protects the repair intermediates to maximize radiation resistance.[13]
Peptide delivery vehicle for biotechnology
Halobacterium sp. NRC-1 is also playing a significant role in the advancement of immunological biotechnology. Recent experimental findings show that recombinant gas vesicles from a mutant strain of Halobacterium sp. NRC-1 have the potential to serve as antigen display/delivery systems. DNA segments of an SIV (simian immunodeficiency virus) gene were inserted into a specific site on a GvpC gene (gas vesicle protein C) in the Halobacterium. The gene was successfully taken up by the microorganism and recombination had been accomplished. Subsequently, the gas vesicles began to express the new recombinant/pathogenic proteins at their surface. Mice were then immunized with these antigen presenting SIV recombinant gas vesicles. Antibody production was monitored, and an increased level of humoral response was observed. After a 12 week period, a booster shot was given. 43 weeks after the booster shot was administered, an elevated number of antibodies still remained and was recorded. Research showed that this increased production of antibodies was long lived even in the absence of an adjuvant, and immunologic memory had been proven. These results indicate that the gas vesicles of Halobacterium sp. NRC-1 show great potential in their possible roles as immunizing agents.[14]
References
- ↑ Woese, C R (1994-03). "There must be a prokaryote somewhere: microbiology's search for itself". Microbiological Reviews 58 (1): 1-9. ISSN 0146-0749. Retrieved on 2009-04-28.
- ↑ Waggoner, Ben; Brian R. Speer (20 April 2001). Introduction to the Archaea: Life's extremists…. University of California Museum of Paleontology. Retrieved on 8 November 2013.
- ↑ McCready, Shirley; Jochen Muller, Ivan Boubriak, Brian Berquist, Wooi Ng, Shiladitya DasSarma (2005). "UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1". Saline Systems 1 (1): 3. DOI:10.1186/1746-1448-1-3. ISSN 1746-1448. Retrieved on 2009-04-18. Research Blogging.
- ↑ 4.0 4.1 4.2 4.3 4.4 Ng, Wailap Victor, et al. (2000-10-24). "Genome sequence of Halobacterium species NRC-1". Proceedings of the National Academy of Sciences of the United States of America 97 (22): 12176-12181. DOI:- 97 VL - 97. Retrieved on 2009-04-18. - 97 Research Blogging.
- ↑ Kottemann, Molly; Adrienne Kish, Chika Iloanusi, Sarah Bjork, Jocelyne DiRuggiero (2005-06-01). "Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation". Extremophiles 9 (3): 219-227. DOI:10.1007/s00792-005-0437-4. Retrieved on 2009-04-28. Research Blogging.
- ↑ 6.0 6.1 6.2 DasSarma, Shiladitya; Brian Berquist, James Coker, Priya DasSarma, Jochen Muller (2006). "Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1". Saline Systems 2 (1): 3. DOI:10.1186/1746-1448-2-3. ISSN 1746-1448. Retrieved on 2009-04-18. Research Blogging.
- ↑ 7.0 7.1 Soppa, Jorg (2006-03-01). "From genomes to function: haloarchaea as model organisms". Microbiology 152 (3): 585-590. DOI:10.1099/mic.0.28504-0. Retrieved on 2009-04-18. Research Blogging.
- ↑ Kennedy, S P; W V Ng, S L Salzberg, L Hood, S DasSarma (2001-10). "Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence". Genome Research 11 (10): 1641-50. DOI:10.1101/gr.190201. ISSN 1088-9051. Retrieved on 2009-04-18. Research Blogging.
- ↑ Müller, Jochen A.; Shiladitya DasSarma (2005-03). "Genomic Analysis of Anaerobic Respiration in the Archaeon Halobacterium sp. Strain NRC-1: Dimethyl Sulfoxide and Trimethylamine N-Oxide as Terminal Electron Acceptors". Journal of Bacteriology 187 (5): 1659–1667. DOI:10.1128/JB.187.5.1659-1667.2005. Retrieved on 2009-04-18. Research Blogging.
- ↑ Coker, James; Priya DasSarma, Jeffrey Kumar, Jochen Muller, Shiladitya DasSarma (2007). "Transcriptional profiling of the model Archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature". Saline Systems 3 (1): 6. DOI:10.1186/1746-1448-3-6. ISSN 1746-1448. Retrieved on 2009-04-18. Research Blogging.
- ↑ Oesterhelt, D (1998-08). "The structure and mechanism of the family of retinal proteins from halophilic archaea". Current Opinion in Structural Biology 8 (4): 489-500. ISSN 0959-440X. Retrieved on 2009-04-26.
- ↑ 12.0 12.1 Weidler, Gerhard (2004-03-01), Survival and growth of Halobacterium sp. NRC-1 following incubation at -15°C, freezing or freeze-drying, and the protective effect of cations, Third European Workshop on Exo-Astrobiology, vol. 545, at 311-312. Retrieved on 2009-04-18
- ↑ Linda C. DeVeaux; Jochen A. Müller, Jonathon Smith, Jill Petrisko, Douglas P. Wells, Shiladitya DasSarma (2007). "Extremely Radiation-Resistant Mutants of a Halophilic Archaeon with Increased Single-Stranded DNA-Binding Protein (RPA) Gene Expression". Radiation Research 168 (4): 507-514. DOI:10.1667/RR0935.1. Retrieved on 2009-05-10. Research Blogging.
- ↑ Sremac, Marinko; Elizabeth S Stuart (2008). "Recombinant gas vesicles from Halobacterium sp. displaying SIV peptides demonstrate biotechnology potential as a pathogen peptide delivery vehicle". BMC Biotechnology 8: 9. DOI:10.1186/1472-6750-8-9. Retrieved on 2009-04-30. Research Blogging.